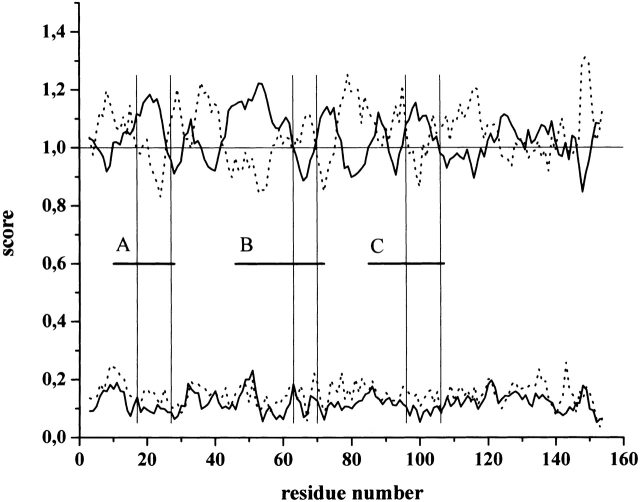
Figure 5.
α-Helicity is preferred within subdomains A and C of calpastatin. All known calpastatin sequences have been downloaded from the Swiss-Prot database; their individual inhibitory domains have been separated and subjected to secondary structure prediction as given in the Materials and Methods section. Inhibitory domains were then aligned by ClustalW to allow the calculation of averages of secondary structure propensity values obtained; where gaps in one or more sequences were generated by the alignment, averaging was done for the remaining sequences. The average for α-helix (solid line) and β-turn (dotted line) is shown for the region encompassing the conserved subdomains A, B, and C marked by thick horizontal lines. For clarity, the significantly lower β-sheet values (~0.8) are not shown. Standard deviation values are seen at the bottom of the figure. Pairs of thin vertical lines mark the regions within each subdomain that are probably in direct contact with calpain in the calpastatin–calpain complex (see text for details).