Abstract
Staphostatins are the endogenous inhibitors of the major secreted cysteine proteases of Staphylococcus aureus, the staphopains. Here, we present the 1.4 Å crystal structure of staphostatin B and show that the fold can be described as a fully closed, highly sheared eight-stranded β-barrel. Thus, staphostatin B is related to β-barrel domains that are involved in the inhibition or regulation of proteases of various catalytic types and to the superfamily of lipocalins/cytosolic fatty acid binding proteins. Unexpectedly for a cysteine protease inhibitor, staphostatin B is not significantly similar to cystatins.
Keywords: β-barrel, cysteine protease inhibitor, SspC, staphostatin
Staphopain B is a secreted protease from the human pathogen Staphylococcus aureus, and has been considered a bacterial virulence factor (Dubin 2002). It belongs to the papain superfamily of cysteine proteases and is encoded in the ssp (staphylococcal serine protease) operon that contains open reading frames (ORFs) for the serine protease V8, the cysteine protease (prepro)staphopain B, and a short ORF termed sspC (Massimi et al. 2002).
Recent work demonstrated that sspC encodes a specific inhibitor of staphopain B (Massimi et al. 2002; Rzychon et al. 2003), and this inhibitor was therefore named staphostatin B (Rzychon et al. 2003). In this communication, we use the staphopain-staphostatin nomenclature. In vitro, staphostatin B has high affinity to staphopain B and inhibits the protease in a 1:1 stoichiometric ratio (Rzychon et al. 2003). In vivo, the lack of an export signal in staphostatin B and the experimentally demonstrated intracellular localization of the protein suggest a role in the protection of S. aureus cells from accidental premature staphopain activation in the cytosol (Rzychon et al. 2003).
As of this writing, the number of known proteins with clear sequence similarity to staphostatin B is rather limited. S. aureus itself contains a staphostatin B homolog in the scp operon that also contains the gene for staphopain A. BLAST (Altschul et al. 1990) searches identified homologs from S. epidermidis, S. warneri, and Clostridium perfringens, but failed to identify reliable clues in the sequence that would have pointed us to a particular class of cysteine protease inhibitors. Thus, we decided to address the question experimentally and solve the crystal structure of staphostatin B.
Results
Staphostatins are β-barrels
The staphostatin B structure was solved as described in Materials and Methods. Data collection and refinement parameters are presented in Table 1. Unexpectedly, staphostatin B turned out to be an eight-stranded mixed β-barrel, with a deviation from the up-down topology of canonical β-barrels in the N-terminal part of the molecule (Fig. 1 ▶). The unwrapped β-barrel is best thought of as a combination of a mixed three-stranded β-sheet and an antiparallel five-stranded sheet that are connected to the complete barrel through a total of six hydrogen bonds only.
Table 1.
Data collection and refinement statistics
| Data collection statistics | Refinement statistics | ||
| Space group | P212121 | R-factor (%) | 20.2 |
| a (Å) | 34.20 | R-free (%) | 22.6 |
| b (Å) | 77.05 | ||
| c (Å) | 101.27 | Rmsd bond distance (Å) | 0.02 |
| Independent reflections | 50616 | Rmsd angles (°) | 1.6 |
| Resolution (Å) | 1.4 | Ramachandran core (%) | 91.7 |
| Completeness (%) | 99 | Ramachandran allowed (%) | 8.3 |
| Rsym (%) (last shell in brackets) | 4.8 (18.8) | Ramachandran gen. all. & disallowed (%) | 0 |
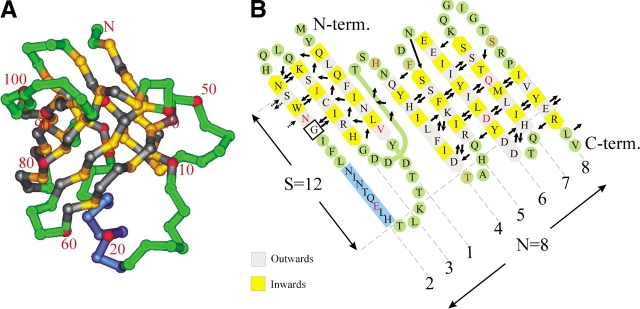
Figure 1.
(A) Cα trace of staphostatin B from the side. Residues in β-strands are yellow and gray, depending on whether they face the inside or outside of the barrel, respectively. Loop regions and turns are green, and the helix is dark blue. Every tenth residue is labeled and marked red. (B) Main chain hydrogen bonding arrangement in staphostatin B as assigned with the program O (Jones et al. 1991). Only hydrogen bonds to or from residues within β-strands are represented and are drawn as arrows pointing from donor to acceptor. Secondary structure assignment was done with dssp (Kabsch and Sander 1983), except for strand β6. Note that for consistency with A, the unwrapped barrel is seen from what was “inside,” resulting in the unconventional slant of β-strands to the left.
The N-terminal part of the molecule is a class 1 Ψ-loop (Hutchinson and Thornton 1990), a rare arrangement with a −2 connection between strands according to the Richardson nomenclature (Richardson 1977). Hydrogen bonds between parallel strands β1 and β3 are exposed to solvent and could thus be unstable (Richardson 1977). Stability may be provided by the +2x crossover connection that links the three-stranded sheet to the five-stranded β-sheet and runs effectively around strand β1. In contrast to the rather unusual fold in the N-terminal part of staphostatin B, the C-terminal part is perfectly canonical, with nearest neighbor (+1) connections between adjacent strands (Fig. 1 ▶).
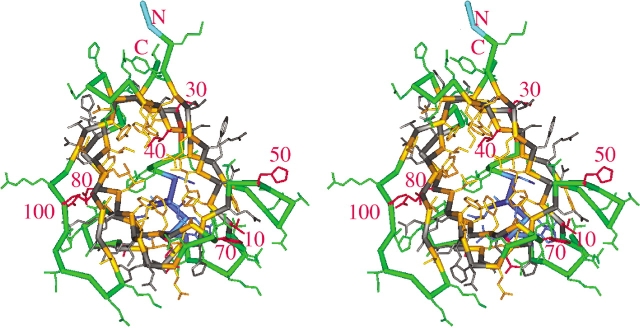
In the terminology of Murzin et al. (1994b), the barrel that results from joining the N-terminal Ψ-loop to the five-stranded sheet at the C terminus through hydrogen bonds on both sides can be described as fully closed. With strand number N=8 and shear number S=12 (Fig. 1 ▶), it obeys the equation S~N+4.2 for barrels of least strain rather than the rules derived from considerations of tight packing of side chains in the barrel interior (Murzin et al. 1994a). For fixed N, the diameter of the barrel increases with shear number S. Thus, the high shear number of staphostatin B is consistent with the presence of a large number of very bulky side chains in the interior of the barrel, especially at the “bottom” of the barrel (Fig. 2 ▶). As expected for barrels with N < S < 2N, the staphostatin B barrel appears flattened and has a cross-section that is far from circular (Fig. 2 ▶).
Figure 2.
Stereo Cα trace of staphostatin B (bold) with side chains (thin lines), looking from the “top” into the eight-stranded β-barrel. The color scheme is as in Figure 1 ▶. The two residues at the N terminus that are left over from the thrombin cleavage step are light blue and are shown without side chains.
Deviations from the canonical β-sheet geometry are rare in staphostatin B, but can be analyzed in detail thanks to the high resolution of our structure; they are presented in diagrammatic form in Figure 1B ▶. In agreement with general preferences (Richardson et al. 1978), β-bulges, the most common disruptions of regular β-sheet geometry, occur only between antiparallel strands and within closely spaced pairs of hydrogen bonds (Fig. 1B ▶). Only one β-bulge occurs away from the loop regions. This “classic” (Chan et al. 1993) β-bulge involves residues L56 and F57 on strand β4 and K66 on strand β5, and introduces a sharp bend into strand β4 (Figs. 1A ▶, 2 ▶). In two instances, hydrogen bonding from one residue to two residues on adjacent strands occurs within the context of four-residue loops. The loop between strands β2 and β3 has the typical “erect hooded cobra” appearance of canonical four-residue loops and conforms to the standard geometry described by Sibanda and Thornton (1985). The second four-residue loop in the structure consists of residues T60, A61, H62, and Q63 and has very similar main chain conformation. A type I reverse turn made of loop residues T84 and Q85 links strands β6 and β7. It follows directly after a classic β-bulge that orients both D82 and D83 towards solvent. With the exception of the three loops described above, turns in staphostatin B are generally wide and not conformationally fixed through main chain hydrogen bonds (Fig. 1B ▶).
Staphostatins resemble lipocalins
Automated, quantitative DALI (Holm and Sander 1995) structure comparisons of staphostatin B and all proteins in the Protein Data Bank (PDB) show that high scorers (Z>3.5) fall into two classes (Table 2): They are either linked to proteolysis as peptidase domains, inhibitors, or regulators, or they belong to the large class of lipocalins/cytosolic fatty acid binding proteins.
Table 2.
Result of a DALI search of the PDB with staphostatin B as the query sequence
| Z | PDB | Protein |
| 6.0 | 1ei5 | Domain C, d-aminopeptidase (dap) |
| 3.8 | 1k3b | Exclusion domain, cathepsin C |
| 2.6 | 1avg | Triabin, in complex with thrombin |
| 4.9 | 1hmt | Human muscle fatty acid binding protein |
| 4.7 | 1epa | Retinoic acid binding protein |
| 4.4 | 1bj7 | Dander major allergen |
| 4.2 | 1iw2 | Complement protein C8 gamma |
| 3.8 | 1beb | β-lactoglobulin |
| 3.7 | 1mup | Major urinary protein |
The DALI-score is a measure of structural similarity between two proteins in standard deviations above the statistically expected similarity (Holm and Sander 1995).
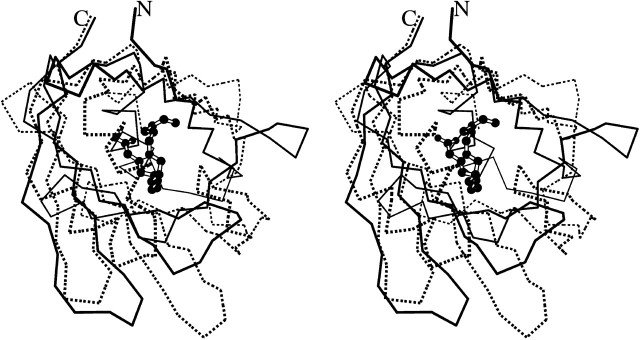
We wondered whether this similarity to lipocalins indicated a secondary, extra function for staphostatins in addition to their properties as protease inhibitors. In a first computational experiment, we searched the staphostatin B structure with VOIDOO (Kleywegt and Jones 1994) for potential internal cavities, but found only cavities of less than 20 Å3 that appear far too small to be functional. In addition, it seems that the highly hydrophobic core of the calyx (or cup)-shaped staphostatin B is not accessible to solvent, at least in the conformation of our crystals. Further evidence against any lipocalin-like properties of staphostatins comes from the superposition with the fatty acid binding protein from human muscle (1HMT), the closest lipocalin-like structural neighbor of staphostatins. Unlike in 1HMT, where the ligand is covered by the “lid” helices of functional lipocalins, a ligand in the analogous position in staphostatins would be entirely exposed to solvent on one side (Fig. 3 ▶).
Figure 3.
Stereo Cα trace of staphostatin B (continuous lines) and of fatty acid binding protein from human muscle (1HMT), the closest lipocalin-like structural neighbor of staphostatins (broken lines). The ligand in 1HMT, stearic acid, is shown in ball-and-stick representation.
Staphostatins versus Human Tear Lipocalin
The protease inhibitory properties of staphostatins have precedence in the discovery of protease inhibitory activity in the lipocalin von Ebner’s gland protein/Human Tear Lipocalin (HTL). On the basis of sequence and mutagenesis data, it has been predicted that HTL forms a regular, strictly antiparallel eight-stranded β-barrel (Fig. 1 ▶ in van’t Hof et al. 1997) and uses three cystatin-related but contiguous motifs within or close to the flexible N terminus of HTL for protease inhibition (van’t Hof et al. 1997; Wojnar et al. 2001). Although the N-terminal part of staphostatins has a Ψ-loop motif instead of the regular up-down topology of lipocalins, we decided to test experimentally whether the N terminus of staphostatin B is required for protease inhibition.
In a first experiment, we used our glutathione-S-transferase (GST)-staphostatin B fusion protein, an intermediate in the purification, for activity assays. We found full inhibitory activity, with no influence from the N-terminal GST-domain. We next deleted the five most N-terminal residues of the wild-type sequence and again found activity in the mutant, even with GST attached. We thus conclude that the N-terminal residues of staphostatin B are not required for inhibitory activity.
Discussion
Lipocalins in Staphylococcus aureus?
The similarity of staphostatins and lipocalins came as a surprise. Staphylococci, so far the only source of staphostatins, are Gram-positive eubacteria. Thus, according to extensive prior bioinformatics work (Ganfornina et al. 2000; Gutierrez et al. 2000), Staphylococcus should not contain lipocalins. Due to the described differences between the N-terminal parts of staphostatins and lipocalins and due to the lack of a cavity in staphostatin B, our present findings do not challenge this hypothesis, but they do show that the absence of lipocalins in Gram-positive bacteria does not imply the absence of molecules with a lipocalin-like fold.
β-barrels in the regulation of proteolysis
There is a precedence for the involvement of β-barrel structures in the regulation of cysteine, serine, and even metalloproteases. Von Ebner’s gland protein/HTL has been classified as a lipocalin on the basis of genetic data (Blaker et al. 1993) and was reported to inhibit papain-like proteases through the involvement of three cystatin-like sequence motifs at the N terminus of the sequence (van’t Hof et al. 1997; Wojnar et al. 2001). In dipeptidyl dipeptidase I (cathepsin C), the β-barrel “exclusion” domain converts the endopeptidase activity that is normally associated with the papain-fold into an exopeptidase activity (Turk et al. 2001). D-aminopeptidase, a serine protease, contains two domains with a lipocalin-like fold, but the domain that is most similar to staphostatin B acts merely as a spacer (Bompard-Gilles et al. 2000). Triabin is a lipocalin-like inhibitor of the serine protease thrombin and interacts exclusively with the fibrinogen recognition exosite of the protease (Fuentes-Prior et al. 1997). Finally, the Erwinia chrysanthemi metalloprotease inhibitor is an eight-stranded antiparallel β-barrel that inserts its N-terminal residues into the primed sites of its target protease (Baumann et al. 1995). Our unpublished data on the staphopain B–staphostatin B complex suggest that this protease-inhibitor complex is different from all of the above cases.
Materials and methods
Crystallization
Staphostatin B (with residues GS at the N terminus left over from the thrombin cleavage step) was produced recombinantly and assayed for activity as described (Rzychon et al. 2003). Crystals were grown by the vapor diffusion method at room temperature (21°C) in sitting drops by equilibrating a 1:1 mixture of 20 mg/mL staphostatin B in 5 mM Tris, pH 7.5 and reservoir buffer against reservoir buffer that contained 160 mM ammonium sulfate, 80 mM sodium acetate pH 4.6, 20% PEG 4K, 20% glycerol, and 10 mM manganese chloride. Crystals grown from this condition contained two molecules in the asymmetric unit, required no buffer exchange for freezing, and diffracted in-house on a Rigaku rotating anode generator to about 1.8 Å. Data collection on a BW6/DESY, Hamburg, yielded a further improvement of the resolution to 1.4 Å with excellent statistics.
Structure determination
Derivatization was achieved with an overnight soak with 0.5 mg/mL uranyl acetate. Isomorphous differences showed consistent Harker peaks on all three Harker sections, and led to the identification of two heavy atom sites with fractional coordinates (0.680, 0.036, 0.238) and (0.490, 0.396, 0.245). Inclusion of the in-house anomalous signal (about 5 electrons for a fully occupied uranium site at 1.54 Å) led to an interpretable map after solvent flattening for one choice of hand. This map was of sufficient quality for near complete main-chain tracing with ARP/wARP (Perrakis et al. 1999; Morris et al. 2002). After the ARP/wARP procedure, maps were of such exceptional quality that discrepancies between the staphostatin sequence from the V8 strain and strain N315 (I70 → F70, T76 → I76) could be read from the electron density map with confidence. Side chains were put manually into the model, and refinement was done with REFMAC (Collaborative Computational Project Number 4 1994). In the final electron density map, there is clear electron density for all residues except the C-terminal V109. The two molecules in the asymmetric unit are largely similar, but differ in the conformation of the two most C-terminal strands. Data collection and refinement parameters are summarized in Table 1.
Acknowledgments
We thank Hans Bartunik and Gleb Bourenkov for generous allocation of beamtime on BW6/DESY and for assistance during data collection, and Izabela Sabala and Roman Szczepanowski for a critical reading of the manuscript. Access to the modeling facilities in ICM and KBN grants 6P04A08320 and 1789/E-529/SPB/5.PR UE/DZ 600/2002-2005 are gratefully acknowledged. This work was done with financial support from the Commission of the European Communities, specific RTD programme “Quality of Life and Management of Living Resources,” QLRT-2001-01250, “Novel Non-antibiotic Treatment of Staphylococcal Diseases.” Structure factors and coordinates are available from the PDB on publication under accession code 1NYC.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Abbreviations
GST, glutathione-S-transferase
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03247703.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Baumann, U., Bauer, M., Letoffe, S., Delepelaire, P., and Wandersman, C. 1995. Crystal structure of a complex between Serratia marcescens metallo-protease and an inhibitor from Erwinia chrysanthemi. J. Mol. Biol. 248 653–661. [DOI] [PubMed] [Google Scholar]
- Blaker, M., Kock, K., Ahlers, C., Buck, F., and Schmale, H. 1993. Molecular cloning of human von Ebner’s gland protein, a member of the lipocalin superfamily highly expressed in lingual salivary glands. Biochim. Biophys. Acta 1172 131–137. [DOI] [PubMed] [Google Scholar]
- Bompard-Gilles, C., Remaut, H., Villeret, V., Prange, T., Fanuel, L., Delmarcelle, M., Joris, B., Frere, J., and Van Beeumen, J. 2000. Crystal structure of a D-aminopeptidase from Ochrobactrum anthropi, a new member of the ‘penicillin-recognizing enzyme’ family. Structure Fold Des. 8 971–980. [DOI] [PubMed] [Google Scholar]
- Chan, A.W., Hutchinson, E.G., Harris, D., and Thornton, J.M. 1993. Identification, classification, and analysis of β-bulges in proteins. Protein Sci. 2 1574–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4. 1994. The CCP4 Suite: Programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 50 760–763. [DOI] [PubMed] [Google Scholar]
- Dubin, G. 2002. Extracellular proteases of Staphylococcus spp. Biol. Chem. 383 1075–1086. [DOI] [PubMed] [Google Scholar]
- Fuentes-Prior, P., Noeske-Jungblut, C., Donner, P., Schleuning, W.D., Huber, R., and Bode, W. 1997. Structure of the thrombin complex with triabin, a lipocalin-like exosite-binding inhibitor derived from a triatomine bug. Proc. Natl. Acad. Sci. 94 11845–11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganfornina, M.D., Gutierrez, G., Bastiani, M., and Sanchez, D. 2000. A phylogenetic analysis of the lipocalin protein family. Mol. Biol. Evol. 17 114–126. [DOI] [PubMed] [Google Scholar]
- Gutierrez, G., Ganfornina, M.D., and Sanchez, D. 2000. Evolution of the lipocalin family as inferred from a protein sequence phylogeny. Biochim. Biophys. Acta 1482 35–45. [DOI] [PubMed] [Google Scholar]
- Holm, L. and Sander, C. 1995. Dali: A network tool for protein structure comparison. Trends Biochem. Sci. 20 478–480. [DOI] [PubMed] [Google Scholar]
- Hutchinson, E.G. and Thornton, J.M. 1990. HERA—A program to draw schematic diagrams of protein secondary structures. Proteins 8 203–212. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 47 (Pt 2) 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. and Sander, C. 1983. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kleywegt, G.J. and Jones, T.A. 1994. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Cryst. D50 178–185. [DOI] [PubMed] [Google Scholar]
- Massimi, I., Park, E., Rice, K., Müller-Esterl, W., Sauder, D., and McGavin, M.J. 2002. Identification of a novel maturation mechanism and restricted substrate specificity for the SspB cysteine protease of Staphylococcus aureus. J. Biol. Chem. 277 41770–41777. [DOI] [PubMed] [Google Scholar]
- Morris, R.J., Perrakis, A., and Lamzin, V.S. 2002. ARP/wARP’s model-building algorithms. I. The main chain. Acta Crystallogr. D. Biol. Crystallogr. 58 968–975. [DOI] [PubMed] [Google Scholar]
- Murzin, A.G., Lesk, A.M., and Chothia, C. 1994a. Principles determining the structure of β-sheet barrels in proteins. I. A theoretical analysis. J. Mol. Biol. 236 1369–1381. [DOI] [PubMed] [Google Scholar]
- ———. 1994b. Principles determining the structure of β-sheet barrels in proteins. II. The observed structures. J. Mol. Biol. 236 1382–1400. [DOI] [PubMed] [Google Scholar]
- Perrakis, A., Morris, R., and Lamzin, V.S. 1999. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6 458–463. [DOI] [PubMed] [Google Scholar]
- Richardson, J.S. 1977. β-Sheet topology and the relatedness of proteins. Nature 268 495–500. [DOI] [PubMed] [Google Scholar]
- Richardson, J.S., Getzoff, E.D., and Richardson, D.C. 1978. The β bulge: A common small unit of nonrepetitive protein structure. Proc. Natl. Acad. Sci. 75 2574–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzychon, M., Sabat, A., Kosowska, K., Potempa, J., and Dubin, A. 2003. Staphostatins: An expanding new group of proteinase inhibitors with a unique specificity for the regulation of staphopains, Staphylococcus spp. cysteine proteinases. Mol. Microbiol. (in press). [DOI] [PubMed]
- Sibanda, B.L., and Thornton, J.M. 1985. β-hairpin families in globular proteins. Nature 316 170–174. [DOI] [PubMed] [Google Scholar]
- Turk, D., Janjic, V., Stern, I., Podobnik, M., Lamba, D., Dahl, S.W., Lauritzen, C., Pedersen, J., Turk, V., and Turk, B. 2001. Structure of human dipeptidyl peptidase I (cathepsin C): Exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases. EMBO J. 20 6570–6582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van’t Hof, W., Blankenvoorde, M.F., Veerman, E.C., and Amerongen, A.V. 1997. The salivary lipocalin von Ebner’s gland protein is a cysteine proteinase inhibitor. J Biol. Chem. 272 1837–1841. [DOI] [PubMed] [Google Scholar]
- Wojnar, P., van’t Hof, W., Merschak, P., Lechner, M., and Redl, B. 2001. The N-terminal part of recombinant human tear lipocalin/von Ebner’s gland protein confers cysteine proteinase inhibition depending on the presence of the entire cystatin-like sequence motifs. Biol. Chem. 382 1515–1520. [DOI] [PubMed] [Google Scholar]