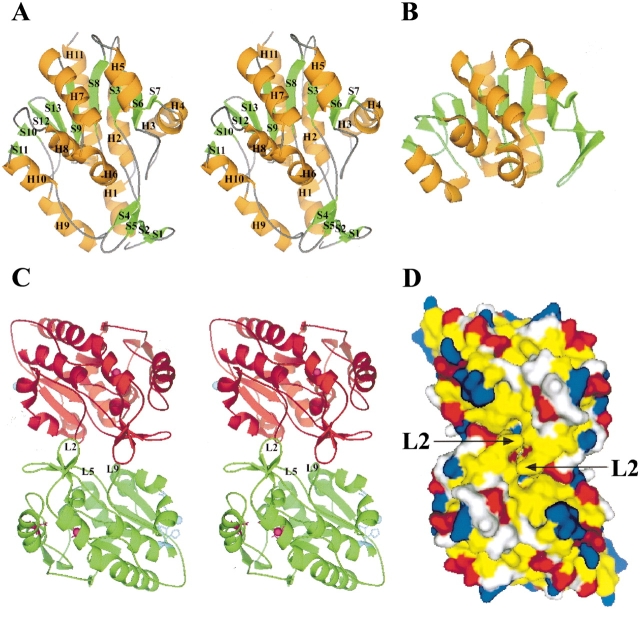
Figure 2.
Overall structure of YedU and structural comparison with protease PH1704. (A) Stereoview of the YedU monomer. H, helix; S, strand; orange and green code for helix and strand, respectively. The upper part of the structure is a sandwich domain (DOM1) composed of a central seven-strand β-sheet flanked by helices. Below the sandwich domain, there is a smaller domain (DOM2) composed of an antiparallel β-sheet (S1↑-S2↓-S5↓-S4↑) and two helices (H1 and H9). Helices H3 and H4 at the right side of the sandwich domain are different from the corresponding region of (B), the structure of PH1704 (Du et al. 2000; PDB acc. code 1G2I), a member of the PfpI cysteine protease family. (C) Stereoview of the YedU homodimer looking down the dimer twofold axis; subunits A and A′ are in green and red, respectively. In subunit A, the β-turn L2 and loops L5 and L9 on the interface are labeled. The potential catalytic triad residues are shown as ball-and-stick in magenta, and the residues coordinating zinc(II) ion are shown as ball-and-stick in cyan. In subunit A′, atom γS of Cys184 and the zinc(II) ion are shown as spheres in magenta and cyan, respectively. (D) The molecular surface of YedU homodimer looking down the molecular twofold axis. Hydrophobic residues (Pro, Ala, Val, Leu, Ile, Met, Phe, Tyr, and Trp) are yellow; positively charged residues (His, Lys, and Arg) are blue; negatively charged residues (Asp and Glu) are red; all other residues are white. Arrows point to the β-turn L2.