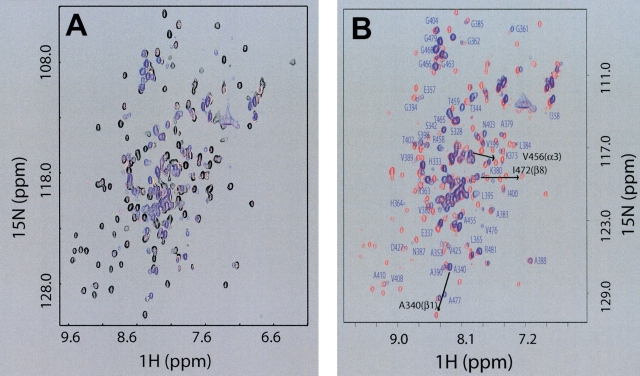
Figure 1.
Interaction with dsDNA causes a reversible local unfolding of d2ΔNS3h. (A) Overlay of 2D 15N-HSQC NMR spectra of uniformly [15N]-labeled d2ΔNS3h acquired at different HSD2/[15N]d2ΔNS3h ratios: 0.0 (black), 0.4 (magenta), 0.62 (yellow), 1.32 (blue). (B) Overlay of 2D 15N-HSQC NMR spectra of purified [15N]d2ΔNS3h–HSD2 complex (blue) and after the dissociation of the complex in the presence of 0.3 M NaCl (red). Isolated peaks in the 15N-HSQC spectrum of purified [15N]d2ΔNS3h–HSD2 complex (blue) are labeled with the assigned one-letter amino acid code and residue number. Labels of additional peaks in crowded regions that have been assigned were omitted for clarity. Also indicated with an arrow and black labels are the chemical shift changes for three peaks, one within each of the three secondary structure elements (β1, α3, β8) that undergo transition from the “unfolded” state (blue: random coil chemical shift in the d2ΔNS3h–HSD2 complex) to the “folded” state (red: dissociated d2ΔNS3h–HSD2 complex upon addition of salt), to further illustrate the localized unfolding of these regions of d2ΔNS3h directly on the dsDNA complex (see text).