Figure 3.
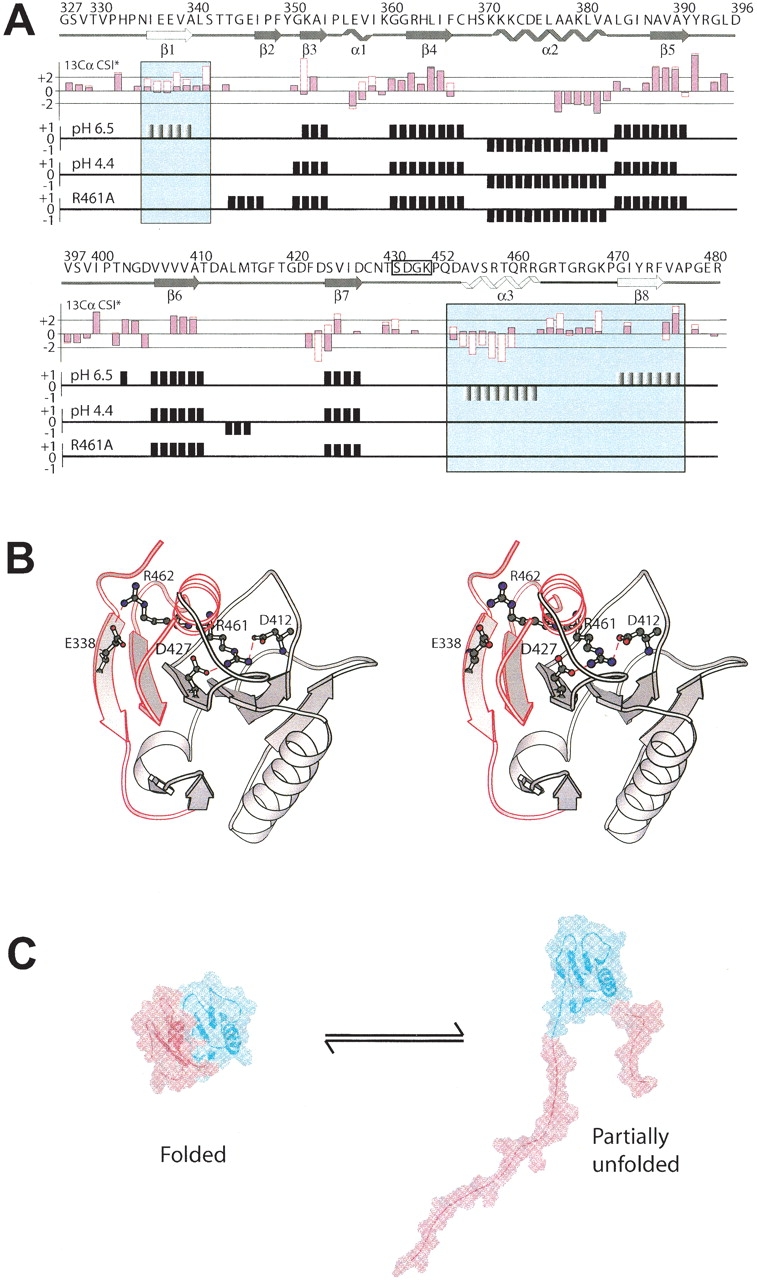
Interaction with dsDNA, lowering the pH to 4.4, or introducing an R461A point mutation all induce a local unfolding of the secondary structure elements β1, β8, and α3 (motif VI) in d2ΔNS3h. (A) Top panel: The primary sequence and the secondary structure of wild-type d2ΔNS3h, as observed in the solution structure at pH 6.5 (Liu et al. 2001) is indicated above the CSI data with the secondary structure elements β1, β8, and α3 highlighted in the panels. Second panel: Comparison of the secondary 13Cα CSI data between d2ΔNS3h–HSD2 complex (empty red boxes) and wild-type d2ΔNS3h (pink filled black boxes) at pH 6.5 for residues whose 13Cα could be assigned in the complex. CSI data of wild-type d2ΔNS3h at pH 6.5 (third panel) and pH 4.4 (fourth panel), and mutant d2ΔNS3h–R461A (fifth panel). (B) Ribbon structure of wild-type d2ΔNS3h at pH 6.5 (Liu et al. 2001) with the side chains of Glu 338, Asp 412, Asp 427, Arg 461, and Arg 462 indicated and labeled. The region which undergoes an order-to-disorder transition is highlighted in red and labeled (β1, β8, and α3). (C) Interaction with dsDNA, lowering the pH to 4.4, or introducing an R461A point mutation all cause the localized unfolding of the region highlighted in red.
