Figure 5.
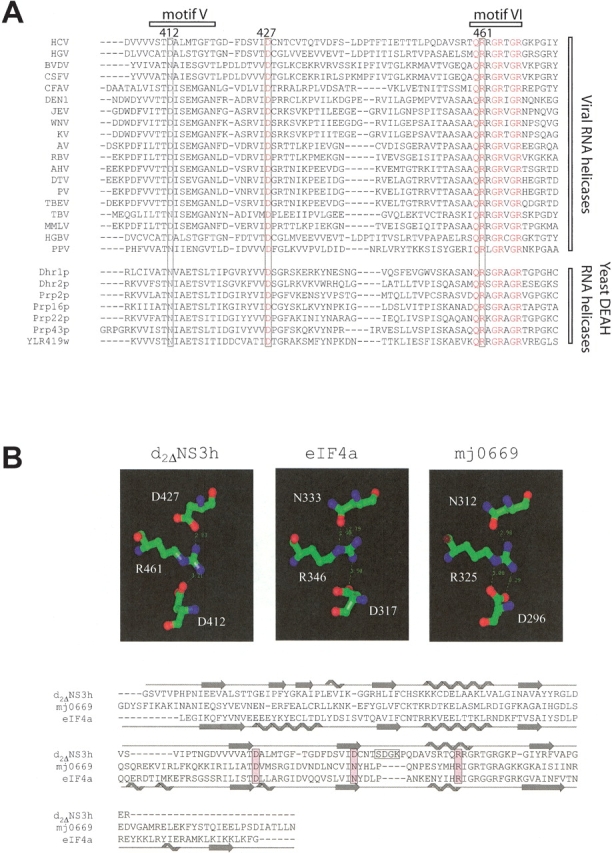
The Arg 461–Asp 412/Asp 427 interactions are conserved both on a sequence and structural level. (A) Sequence alignment of several viral SF2 helicases, which are homologous to the HCV NS3 helicase (HCV). These proteins include the homologous NS3 proteins of hepatitis G virus (HGV), two pestiviruses (bovine viral diarrhea virus [BVDV] and classical swine fever virus [CSFV]), and 14 flaviviruses (cell fusing agent virus [CFAV], dengue virus type 1 [DEN1], Japanese encephalitis virus [JEV], West Nile virus [WNV], kunjin virus [KV], apoi virus [AV], Rio Bravo virus [RBV], alkhurma virus [AHV], deer tick virus [DTV], powassan virus [PV], tick-borne encephalitis virus [TBEV], Tamana bat virus [TBV], Montana myotis leukoencephalitis virus [MMLV], and marmoset hepatitis GB virus [HGBV]). Also shown is the sequence of the CI helicase of a plant potyvirus, plum pox virus (PPV), and those of seven DEAH-box proteins from the yeast Saccharomyces cerevisiae, which are classified as RNA helicases (http://www.expasy.ch/linder/helicases_list.html). (B) Structural comparison of three SF2 helicases (d2ΔNS3h; mj0669, a DEAD-box putative RNA helicase from Methanococcus jannaschi; and eIF4a [eukaryotic translation initiation factor 4A] from yeast, a DEAD-box RNA helicase) illustrating the conservation of the Arg 461–Asp 412/Asp 427 interactions of the HCV helicase in eIF4a and mj0669. The secondary structure elements are indicated above and below the sequence alignment for d2ΔNS3h and eIF4a, respectively.
