Abstract
We report NMR assignments and solution structure of the 71-residue 30S ribosomal protein S28E from the archaean Pyrococcus horikoshii, target JR19 of the Northeast Structural Genomics Consortium. The structure, determined rapidly with the aid of automated backbone resonance assignment (AutoAssign) and automated structure determination (AutoStructure) software, is characterized by a four-stranded β-sheet with a classic Greek-key topology and an oligonucleotide/oligosaccharide β-barrel (OB) fold. The electrostatic surface of S28E exhibits positive and negative patches on opposite sides, the former constituting a putative binding site for RNA. The 13 C-terminal residues of the protein contain a consensus sequence motif constituting the signature of the S28E protein family. Surprisingly, this C-terminal segment is unstructured in solution.
Keywords: Ribosomal protein, Greek-key motif, NMR structure, Northeast Structural Genomics Consortium
The field of structural genomics aims at elucidating the structures of representative proteins from sequence families to provide more complete coverage of protein fold space and at spurring advances in bioinformatic, biotechnological, and biophysical techniques that will make high-throughput protein production and structure determination feasible (Montelione et al. 2000; Burley and Bonanno 2002). The Northeast Structural Genomics Consortium (NESG; www.nesg.org), a pilot project funded by the National Institutes of Health Protein Structure Initiative, is focused on sequence-based clusters of eukaryotic protein domain families. Our efforts target one or more members of protein domain families from eukaryotic proteomes (Liu et al. 2003) and their homologs in bacteria and archaea, particularly proteins with low sequence similarity to proteins of known structure.
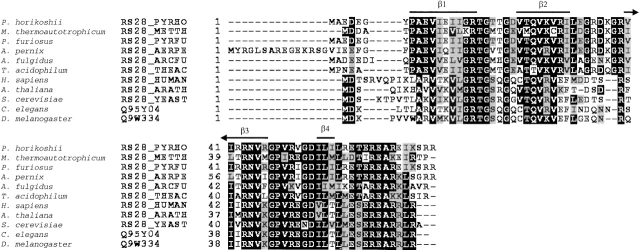
The 71-residue S28E protein from the 30S ribosomal unit of Pyrococcus horikoshii (SWISS-PROT ID: RS28_PYRHO, Boeckmann et al. 2003; NESG target i.d. JR19; Wunderlich et al. 2003) is a member of a conserved protein domain family ubiquitous in archaea and eukaryotes. In spite of an abundance of structural information for ribosomal proteins from recent X-ray crystal structures of a bacterial 30S subunit (Wimberley et al. 2000), an archaeal 50S subunit (Ban et al. 2000), and an intact bacterial ribosome (Yusupov et al. 2001), little is known about the role of the S28E family in archaeal and eukaryotic ribosomes. Moreover, no three-dimensional structure of any member of the S28E domain family has yet been described in the literature. The multiple sequence alignment of S28E with homologs from other model organisms that constitute the NESG cluster of targets for this protein family (http://cubic.bioc.columbia.edu/cgi-bin/var/database/pep/getPepEntry.cgi?clusterId=18021 [Liu et al. 2003]), including that from Methanobacterium thermoautotrophicum (NESG target TT744; 64% sequence identity to S28E), is shown in Figure 1 ▶. In this paper, we describe the NMR solution structure of S28E, discuss the insights gained from the structure into the function of the S28E family of ribosomal proteins, and compare it to the solution NMR structure of M. thermoautotrophicum S28E described in the accompanying paper (Wu et al. 2003).
Figure 1.
Multiple sequence alignment of the S28E protein from Pyrococcus horikoshii (NESG target JR19) with proteins (SWISS-PROT ID shown) from NESG cluster 18021 (Liu et al. 2003; http://cubic.bioc.columbia.edu/PEP), aligned using Clustal X (Jeanmougin et al. 1998). Identical and similar residues are box shaded in black and gray, respectively. Regions of secondary structure observed in the solution NMR structure of S28E are indicated above the sequence.
Results and Discussion
Resonance assignments
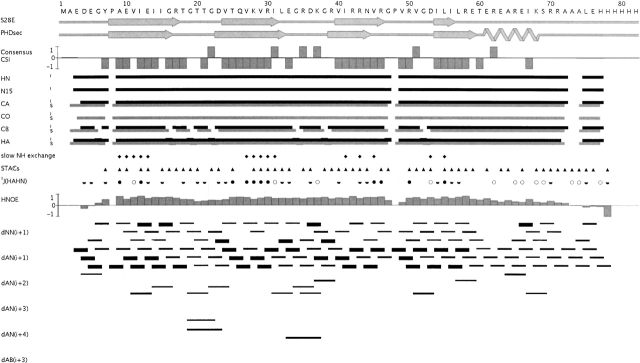
Using the automated assignment program AutoAssign (Zimmerman et al. 1997; Moseley et al. 2001) with a three-rung (Cα, Cβ, Hα) matching strategy for establishing sequential spin system relationships, together with spin-system-type assignment constraints (STACs; Zheng et al. 2003), we obtained ~98% complete assignment of backbone nuclei of S28E, excluding the C-terminal tag (HN-N: 67/68; C′: 69/71; Cα: 70/71; Hα: 78/79). The resulting AutoAssign connectivity map for S28E is shown in Figure 2 ▶. The assignments from AutoAssign were manually extended into the side chains, yielding ~92% complete side chain assignment, excluding the C-terminal tag: (Cβ: 62/63; Cγ: 68/70; Cδ: 34/35; Cɛ: 5/6; Hβ: 100/102; Hγ: 104/106; Hδ: 54/54; Hɛ: 10/24; Nɛ: 1/14). In addition, stereospecific isopropyl methyl assignments were obtained for all valines and leucines in the protein (12 in total).
Figure 2.
AutoAssign connectivity map for S28E. Intraresidue (i) and sequential (s) connectivities are shown as horizontal black and gray lines, respectively. Resonances of residues A73 and A74, as well as H78 through H82 in the C-terminal tag, could not be unambiguously assigned due to degeneracy. Sequential spin-system type assignment constraints (STACs) used in the AutoAssign calculations are represented by filled triangles. 3J(HN-Hα) values are depicted as open circles (<6 Hz), half-filled circles (6 ≤ J ≤ 8 Hz), and filled circles (>8 Hz). Residues with slowly exchanging backbone amide protons are represented by filled diamonds. Interresidue NOE connectivities identified by AutoStructure calculations are shown as thin, medium, and thick black lines, corresponding to weak, medium, and strong NOE interactions. Bar graphs of the consensus CSI (Wishart and Sykes 1994) and 1H-15N heteronuclear NOE data are shown. Secondary structural elements in the final S28E structure and predicted by PHD (Rost 1996) are also shown.
Description of the three-dimensional structure
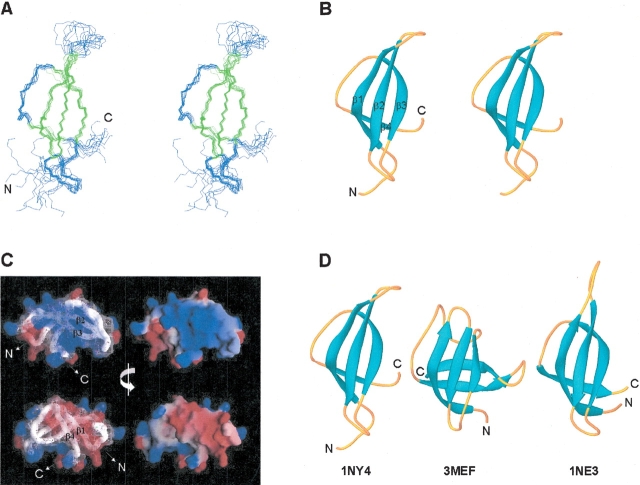
The three-dimensional solution structure of S28E was determined by automated analysis of NMR data using the programs AutoStructure (Huang 2001; Huang et al. 2003; Zheng et al. 2003) and DYANA (Güntert et al. 1997). The solution structure is represented by the 10 conformers that best fit the resulting NMR constraint data. The backbone atoms of the superimposed ensemble, together with ribbon and electrostatic surface potential diagrams of a representative structure, are shown in Figure 3A–C ▶, and structural statistics are listed in Table 1. The structure of S28E is comprised of a Greek-key motif with four β-strands, encompassing residues Pro 8–Arg 17 (which includes β-bulges at Ile 12 and Ile 15), Val 24–Ile 31, Val 40–Arg 46, and Ile 54–Ile 56. These strands interact to form two antiparallel β-sheets, one four-stranded (3–2–1–4) and one three-stranded (3–2–1) β-sheet, packing into an overall β-barrel fold (Fig. 3A,B ▶). This topology is strongly corroborated by several lines of spectral evidence, including the pattern of sequential and medium range NOEs, the slow amide exchange pattern, and local backbone dynamical information indicated by 1H-15N heteronuclear NOE data (Fig. 2 ▶). Both β-bulges in β-strand 1 are characterized by strong sequential HN-HN NOEs, and a small 3J(HN-Hα) scalar coupling constant was observed for Val 11. The electrostatic surface potential (Fig. 3C ▶) reveals that the molecule has two distinct charged surfaces: a positively charged surface formed by basic residues from β-strands 2 and 3, and a negatively charged surface due to a cluster of acidic residues from β-strands 1 and 4 and the loops between β-strands 2 and 3, and between β-strands 3 and 4. Several of these charged residues, including Lys 28, Arg 30, Arg 39, Arg 43, Glu 10, and Asp 53, are highly conserved across the S28E family (Fig. 1 ▶). Interestingly, the C-terminal ~13 residues of S28E (plus the C-tag) are essentially disordered in the structure, and there is little spectroscopic evidence (i.e., NOE patterns, CSI, etc.) for the α-helix that is predicted (Rost 1996) in this region of the molecule (Fig. 2 ▶). Conformational flexibility in this C-terminal region of S28E is also indicated by 1H-15N heteronuclear NOE data (Fig. 2 ▶). Small 3J(HN-Hα) scalar coupling constants were measured throughout the C-terminal polypeptide segment, suggesting the possible formation of a transient α-helix. Interestingly, this unstructured C-terminal region features a PROSITE (Falquet et al. 2002) sequence, E-[S/T]-E-R-E-A-R-x-[L/I], that is a signature of the S28E family of proteins.
Figure 3.
NMR solution structure of S28E determined using AutoStructure and DYANA. For the purposes of display, the disordered C-terminal segment and His-tag have been omitted, and only residues 1 through 60 are shown. (A) Stereoview showing the backbone atom superposition of 10 conformers representing the solution structure; β-strand elements are shown in green. (B) Stereoview of a ribbon representation of a representative conformer (lowest DYANA target function) from the ensemble. The four β-strands comprising the Greek-key motif are labeled. (C) Electrostatic potential surfaces (Nicholls et al. 1991) showing the “positive” (blue) and “negative” (red) faces of S28E. (D) Ribbon diagrams of representative models from the solution structures of P. horikoshii S28E (1NY4; residues 1 to 60), E. coli CspA (3MEF), and M. thermoautotrophicum S28E (1NE3; residues 1 to 58). The superpositions (A) were made using the program MOLMOL (Koradi et al. 1996) and ribbon diagrams (B and D), were generated by the program Ribbons (Carson 1991).
Table 1.
Summary of NMR data and structural statistics for S28E
| Completeness of resonance assignmentsa | |||
| Assignable backbone (%) | 98 | ||
| Assignable side chain (%) | 92 | ||
| 3D 15N | 3D 13C | 4D 13C/13C | |
| NOESY spectral datab | |||
| Total number of peaks | 747 | 1462 | 920 |
| Number of “assignable” peaks | 744 | 1352 | 910 |
| Number of peak assignments | 618 | 1198 | 771 |
| Percent assignment (%) | 83 | 89 | 85 |
| Conformationally restricting constraints | |||
| Distance constraints | |||
| Total | 828 | ||
| Intraresidue (i = j) | 121 | ||
| Sequential (|i − j| = 1) | 268 | ||
| Medium range (1 < |i − j| ≤ 5) | 97 | ||
| Long range (|i − j| > 5) | 342 | ||
| Distance constraints per residue | 11.7 | ||
| Dihedral angle constraints | 69 | ||
| Hydrogen bond constraints | |||
| Total | 30 | ||
| Long range (|i − j| > 5) | 28 | ||
| Number of constraints per residue | 13.1 | ||
| Number of long range constraints per residue | 5.2 | ||
| Residual constraint violationsc | |||
| DYANA target function (Å2) | 0.48 ± 0.067 | ||
| Average number of distance violations per structure | |||
| 0.1–0.2 Å | 6.5 | ||
| 0.2–0.5 Å | 2.4 | ||
| >0.5 Å | 0 | ||
| Average r.m.s. distance violation per constraint (Å) | 0.02 | ||
| Maximum distance violation (Å) | 0.28 | ||
| Average number of dihedral angle violations per structure | |||
| 0–10° | 0.7 | ||
| >10° | 0 | ||
| Average r.m.s. dihedral angle violation per constraint (°) | 0.13 | ||
| Maximum dihedral angle violation (°) | 1.30 | ||
| van der Waals violations per structure | |||
| Average van der Waals violation (Å) | 0.10 | ||
| Maximum van der Waals violation (Å) | 0.12 | ||
| r.m.s.d. from average coordinates; ordered residuesd | |||
| Backbone atoms | 0.3 | ||
| Heavy atoms | 1.1 | ||
| Ramachandran plot statistics; ordered residuesd | |||
| Most favored regions (%) | 82.0 | ||
| Additional allowed regions (%) | 18.0 | ||
| Generously allowed (%) | 0.0 | ||
| Disallowed regions (%) | 0.0 | ||
| r.m.s. deviations from ideal geometry | |||
| Bond length (Å) | 0.011 | ||
| Bond angle (°) | 1.7 | ||
Structural statistics were compared for the 10 structures with lowest DYANA target function out of 56 calculated in the final cycle of AutoStructure analysis.
a Computed using AVS software (H.N.B. Moseley, G. Sahota, and G.T. Montelione, in prep.) from the expected number of peaks, excluding highly exchangeable protons (N-terminal, Lys, and Arg amino groups, hydroxyls of Ser, Thr, Tyr), carboxyls of Asp and Glu, and nonprotonated aromatic carbons.
b Peak assignment statistics for the 3D 15N, combined aliphatic and aromatic 3D 13C, and 4D 13C/13C NOESY spectra from the final cycle of AutoStructure. The total number of peaks refer to the raw input to AutoStructure; the number of “assignable” peaks refer to peaks that can be matched to the chemical shift list within the tolerances used; the number of peak assignments represents the total (unambiguous + ambiguous) number of peaks assigned by AutoStructure.
c Average distance violations were calculated using the sum over r−6.
d Ordered residue ranges: 7–17, 24–33, 36–47, 50–56.
Comparison to related structures
A Dali (Holm and Sander 1993) search of the Protein Data Bank (PDB) revealed that the structure of S28E is distantly related to the classic OB-fold (βββαββ) found in many oligosaccharide and nucleic acid binding proteins (Murzin 1993; Arcus 2002), although it lacks two structural elements commonly seen in OB-folds, the α-helix between β-strands 3 and 4 and the fifth β-strand. The closest structural relative in the PDB is the 70-residue CspA cold shock protein from Escherichia coli, whose structure has been solved by both X-ray crystallography (Schindelin et al. 1994; 1MJC; Dali Z score = 4.4) and NMR (Newkirk et al. 1994; Feng et al. 1998; 3MEF). In spite of the very low sequence identity between S28E and CspA (11%), the four β-strands of S28E superimpose well with the first four strands of the OB-fold of CspA, including the β-bulge at Ile 12 in β-strand 1. However, instead of a fifth β-strand folding back to close the β-barrel, as in the CspA structure, the C terminus of S28E juts away from the core of the molecule (Fig. 3D ▶). Like CspA, S28E lacks an α-helical segment between β-strands 3 and 4, typical of other OB-fold proteins. The positively charged surface of S28E is similar to the positively charged nucleic-acid-binding epitope of CspA, formed by conserved basic and surface aromatic residues predominantly from β-strands 1, 2, and 3. However, in contrast to CspA, S28E, and indeed the entire S28E family, is largely devoid of aromatic residues, which play a key role in nucleic acid binding by CspA (Schröder et al. 1995). Other proteins containing OB-fold motifs structurally similar to S28E include the laminin-binding domain of agrin (Stetefeld et al. 2001; 1J7C), the maltose transport protein MalK (Diederichs et al. 2000; 1G29), and the molybdate-binding protein ModG (Delarbre et al. 2001; 1H9J).
The OB-fold is employed for RNA binding by several proteins, including ribosomal proteins S1, S12, and S17 (Draper and Reynoldo 1999; Brodersen et al. 2002). Interestingly, in the crystal structure of the 30S subunit from Thermus thermophilus, ribosomal protein S17 (104 amino acids) features a long C-terminal α-helix following the five-stranded β-barrel, with residues in loops as well as in this α-helix making contacts with specific RNA elements at the interface between the 5′ and central domains of the 30S subunit (Wimberly et al. 2000; Brodersen et al. 2002). The unstructured C-terminal segments of S28E and TT744, which are predicted to form α-helices (Rost 1996), may play a similar role in RNA binding, becoming ordered only upon making specific interactions with rRNA substrate and/or other proteins.
The solution NMR structure of S28E is also very similar to that of the related S28E protein from Methanobacterium thermoautotrophicum (NESG TT744) described in the accompanying paper (Wu et al. 2003); the proteins share 64% sequence identity. The ordered regions of these homologs are highly superimposable, with a pairwise backbone (N, C′, Cα) root-mean-square deviation of 0.72 ± 0.14 Å over the ensembles (Fig. 3D ▶). It is interesting to note that these two solution NMR structures were derived using different automated approaches for determining NOESY cross-peak assignments. In both proteins the highly conserved C-terminal region predicted (Rost 1996) to be α-helical is disordered, and it is reasonable to predict that it may become structured upon binding to other proteins and/or rRNA in the ribosome.
Materials and methods
Sample preparation
The full-length S28E gene (RPS28E) from P. horikoshii was cloned into a pET21d (Novagen) derivative, generating plasmid pJR19–21. The resulting S28E open reading frame contains an additional 11 nonnative residues (AAALEHHHHHH) at the C terminus of the protein. This construct sequence was verified by standard DNA sequence analysis. E. coli BL21 (DE3) pMGK cells, a rare codon-enhanced strain, were transformed with pJR19–21. A single isolate was cultured in MJ9 minimal medium (Jansson et al. 1996), containing 100% 15N-enriched ammonium sulphate and either 5% or 100% uniformly enriched 13C-glucose as the sole nitrogen and carbon sources, respectively. Initial growth was carried out at 37°C until the O.D.600 of the culture reached 1.0 units. The incubation temperature was then decreased to 17°C and protein expression was induced by the addition of isopropyl-β-D-thiogalactopyranoside at a final concentration of 1 mM. Following overnight incubation at 17°C, the cells were harvested by centrifugation.
S28E protein samples were purified using standard protocols. Cell pellets were resuspended in Lysis Buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 5 mM β-mercaptoethanol at pH 8.0) and disrupted by sonication. The resulting lysate was clarified by centrifugation at 26,000g for 45 min at 4°C. The supernatant was loaded onto a Ni-NTA column (Qiagen) and eluted in Lysis Buffer containing 250 mM imidazole. Fractions containing partially purified S28E were pooled and loaded onto a gel filtration column (Sephadex 75, Amersham Pharmacia Biotech), and eluted in 10 mM Tris, 100 mM NaCl, and 5 mM dithiothreitol (DTT) at pH 8.0. Sample purity (>97%) and molecular weight (9718 D) were verified by SDS-PAGE and MALDI-TOF mass spectrometry, respectively. The yield of purified protein was approximately 20 mg/L. Protein samples for NMR spectroscopy were concentrated by ultracentrifugation to 1.0 mM S28E in 20 mM MES, 100 mM NaCl, 5 mM CaCl2, 10 mM DTT (pH 6.5), containing 5% (v/v) 2H2O (unless otherwise indicated).
NMR spectroscopy
NMR spectra were acquired at 20°C on 500, 600, 750, and 800 Varian Inova spectrometers and a 600 Varian Unity spectrometer. Chemical shifts are referenced to external DSS, whereas 13C and 15N chemical shifts were referenced indirectly using the gyromagnetic ratios of 13C:1H (0.251449530) and 15N:1H (0.101329118), respectively. Backbone and side chain resonance assignments were made using the following series of standard triple-resonance experiments (Cavanaugh et al. 1996; Montelione et al. 1999) obtained on U-13C,15N S28E: 2D 15N-1H HSQC, 2D 13C-1H HSQC, 3D HNCO, HNCACB, CBCA(CO)NH, HA(CA)(CO)NH, HA(CA)NH, CBCACO(CA)HA, (H)CC(CO)NH-TOCSY, and H(CC)(CO)NH-TOCSY, along with HCCH-TOCSY and HCCH-COSY spectra recorded on a sample prepared in 100% 2H2O. NOE distance constraints were derived from 3D 15N-edited NOESY (τm = 80 ms), 3D 13C-edited aliphatic and aromatic NOESYs (τm = 80 ms), and 4D 13C/13C -NOESY in 100% 2H2O (τm = 100 ms). Three-bond 3J(HN-Hα) scalar couplings were obtained from the ratio of cross-peak to diagonal intensities in a 3D HNHA (Vuister and Bax 1993). Amide proton exchange rate information was obtained by a 1:6 dilution of 50 μL U-13C,15N S28E with 100% 2H2O and monitoring the decay of the 15N-1H HSQC signal height over time; some amide resonances in the core of the protein were still present after several days. Stereospecific Val and Leu methyl assignments were obtained from a high-resolution nonconstant time 13C-1H HSQC spectrum of 5%-13C, U-15N S28E (Neri et al. 1989). 1H-15N heteronuclear NOEs were obtained on 5%-13C, U-15N S28E using a sensitivity-enhanced version of our previously described 2D heteronuclear NOE experiment (Li and Montelione 1993), with a recycle time of 14 sec to ensure complete relaxation of solvent magnetization. All NMR spectra were processed with NMRPipe 2.1 (Delaglio et al. 1995) and analyzed with Sparky 3.106 (Goddard and Kneller 2002).
Resonance assignments
Backbone HN, N, C′, Cα, Hα, and side chain Cβ resonance assignments were obtained using AutoAssign 1.9 (Zimmerman et al. 1997; Moseley et al. 2001), employing peak lists from the 15N-1H HSQC, HNCO, HNCACB, CBCA(CO)NH, HA(CA)(CO)NH, and HA(CA)NH spectra. Sequential STACs information (Zheng et al. 2003), obtained from 2D (HN-N plane) Gly-phased HA(CACO)NH (Feng et al. 1996) and (H)CC(CO)NH-TOCSY data, were included in the AutoAssign analysis. These assignments were extended into the side chains by manual analysis of (H)CC(CO)NH-TOCSY, H(CC)(CO)NH-TOCSY, HCCH-TOCSY, HCCH-COSY, and 13C-edited NOESY spectra. Resonance assignments were validated using the Assignment Validation Suite (AVS) software package (H.N.B. Moseley, G. Sahota, and G.T. Montelione, in prep.).
Automated structure determination
Structure calculations were performed using the program AutoStructure 1.1.2 (Huang 2001; Huang et al. 2003; Zheng et al. 2003), interfaced with DYANA 1.5 (Güntert et al. 1997). Briefly, AutoStructure performs iterative NOESY assignment and structure calculations using DYANA, XPLOR, or CNS (Brünger et al. 1998) in an automated fashion for a user-defined number of cycles. After generating a reliable initial protein fold based in the first cycle of NOESY spectral analysis, AutoStructure automatically generates distance (NOE), dihedral angle (using the program HYPER; Tejero et al. 1999), and hydrogen-bond constraints in successive cycles. In this case, the input for the AutoStructure program consisted of a resonance assignment list, manually edited peak lists with intensities for the 3D 15N-edited, 3D 13C-edited, and 4D 13C/13C NOESY spectra, 3J(HN-Hα) values, φ,ξ angle constraints (±40° and ±50°, respectively) derived from chemical shift data using the program TALOS (Cornilescu et al. 1999), and slow amide hydrogen exchange data. TALOS dihedral constraints were used only for residues with confidence scores of 10. Tolerances for matching NOESY peaks with resonance assignments within the NOESY assign module of the program were set to ±0.05 ppm for 1H (±0.04 ppm for the directly detected dimension in the 3D NOESY spectra) and ±0.5 ppm for 13C and 15N, and the program automatically handles aliased peaks. The reported ensemble of structures comprises the best 10 of 56 structures from the final cycle of AutoStructure, on the basis of DYANA target function. Structures were analyzed using the programs PDBStat (R. Tejero and G.T. Montelione, unpubl.) and PROCHECK-NMR (Laskowski et al. 1996). The final ensemble of structures (minus the C-terminal tag) and structural constraints, as well as the chemical shift, 3J(HN-Hα) data, and raw fids have been deposited in the Protein Data Bank (PDB ID 1NY4) and BioMagRes Bank (accession number 5691), respectively.
Acknowledgments
We thank C. Arrowsmith, A. Bhattacharrya, H. Moseley, R. Tejero, and B. Wu for helpful discussions. This work was supported by a grant from the Protein Structure Initiative of the NIH (P50 GM62413) and by NSF grant DBI-9904841. The majority of the NMR spectra were acquired in the Environmental Molecular Sciences Laboratory (a national scientific user facility sponsored by the U.S. Department of Energy (DOE) Office of Biological and Environmental Research) located at Pacific Northwest National Laboratory and operated for DOE by Battelle (contract KP130103).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.03359003.
References
- Arcus, V. 2002. OB-fold domains: A snaphot of the evolution of sequence, structure and function. Curr. Opin. Struct. Biol. 12 794–801. [DOI] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289 905–920. [DOI] [PubMed] [Google Scholar]
- Boeckmann, B., Bairoch, A., Apweiler, R., Blatter, M.-C., Estreicher, A., Gasteiger, E., Martin, M.J., Michoud, K., O’Donovan, C., Phan, I., et al. 2003. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 31 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen, D.E., Clemons Jr., W.M., Carter, A.P., Wimberly, B.T., and Ramakrishnan, V. 2002. Crystal structure of the 30S ribosomal subunit from Thermus thermophilus: Structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 316 725–768. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta. Crystallogr. D 54 905–921. [DOI] [PubMed] [Google Scholar]
- Burley, S.K. and Bonanno, J.B. 2002. Structuring the protein universe. Annu. Rev. Genomics Hum. Genet. 3 243–262. [DOI] [PubMed] [Google Scholar]
- Carson, M. 1991. Ribbons 2.0. J. Appl. Crystallogr. 24 958–961. [Google Scholar]
- Cavanaugh, J., Fairbrother, W.J., Palmer III, A.G., and Skelton, N.J. 1996. Protein NMR spectroscopy. Academic Press, San Diego, CA.
- Cornilescu, G., Delaglio, F., and Bax, A. 1999. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13 289–302. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6 277–293. [DOI] [PubMed] [Google Scholar]
- Delarbre, L., Stevenson, C.E., White, D.J., Mitchenall, L.A., Pau, R.N., and Lawson, D.M. 2001. Two crystal structures of the cytoplasmic molybdate-binding protein ModG suggest a novel cooperative binding mechanism and provide insights into ligand-binding specificity. J. Mol. Biol. 308 1063–1079. [DOI] [PubMed] [Google Scholar]
- Diederichs, K., Diez, J., Greller, G., Müller, C., Breed, J., Schnell, C., Vonrhein, C., Boos, W., and Welte, W. 2000. Crystal structure of MalK, the ATPase subunit of the trehalose/maltose ABC transporter of the archaeon Thermococcus litoralis. EMBO J. 19 5951–5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper, D.E. and Reynaldo, L.P. 1999. RNA binding strategies of ribosomal proteins. Nucleic Acids Res. 27 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falquet, L., Pagni, M., Bucher, P., Hulo, N., Sigrist, C.J.A., Hofmann, K., and Bairoch, A. 2002. The PROSITE database, its status in 2002. Nucleic Acids Res. 30 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, W., Rios, C.B., and Montelione, G.T. 1996. Phase labeling of C-H and C-C spin-systems topologies: Application in PFG-HACANH and PFG-HACA(CO)NH triple-resonance experiments for determining backbone resonance assignments in proteins. J. Biomol. NMR 8 98–104. [DOI] [PubMed] [Google Scholar]
- Feng, W., Tejero, R., Zimmerman, D.E., Inouye, M., and Montelione, G.T. 1998. Solution NMR structure and backbone dynamics of the major cold-shock protein (CspA) from Escherichia coli: Evidence for conformational dynamics in the single-stranded RNA-binding site. Biochemistry 37 10881–10896. [DOI] [PubMed] [Google Scholar]
- Goddard, T.D. and Kneller, D.G. 2002. Sparky 3. University of California, San Francisco. http://cgl.ucsf.edu/home/sparky.
- Güntert, P., Mumenthaler, C., and Wüthrich, K. 1997. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273 283–298. [DOI] [PubMed] [Google Scholar]
- Holm, L. and Sander, C. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233 123–128. [DOI] [PubMed] [Google Scholar]
- Huang, Y.J. 2001. “Automated determination of protein structures from NMR data by iterative analysis of self-consistent contact patterns.” Ph.D. thesis, Rutgers University, New Brunswick, NJ.
- Huang, Y.J., Swapna, G.V.T., Rajan, P.K., Ke, H., Xia, B., Shukla, K., Inouye, M., and Montelione, G.T. 2003. Solution NMR structure of ribosome-binding factor-A (RbfA), a cold-shock adaptation protein from Escherichia coli. J. Mol. Biol. 327 521–536. [DOI] [PubMed] [Google Scholar]
- Jansson, M., Li, Y.-C., Jendeberg, L., Anderson, S., Montelione, G.T., and Nilsson, B. 1996. High level production of uniformly 15N- and 13C-enriched fusion proteins in Escherichia coli. J. Biomol. NMR 7 131–141. [DOI] [PubMed] [Google Scholar]
- Jeanmougin, F., Thompson, J.D., Gouy, M., Higgins, D.G., and Gibson, T.J. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23 403–405. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wüthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graphics 14 51–55. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., Rullmann, J.A., MacArthur, M.W., Kaptein, R., and Thornton, J.M. 1996. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8 477–486. [DOI] [PubMed] [Google Scholar]
- Li, Y.-C. and Montelione, G.T. 1993. Solvent saturation-transfer effects in pulsed-field-gradient heteronuclear single-quantum-coherence (PFG-HSQC) spectra of polypeptides and proteins. J. Magn. Reson. Ser. B 101 315–319. [Google Scholar]
- Liu, J., Hegyi, H., Acton, T.B., Montelione, G.T., and Rost, B. 2003. Automatic target selection for structural genomics on eukaryotes. Protein Sci. (in press) [DOI] [PubMed]
- Montelione, G.T., Rios, C.B., Swapna, G.V.T., and Zimmerman, D.E. 1999. NMR pulse sequences and computational approaches for automated analysis of sequence-specific backbone resonance assignments of proteins. In Biological magnetic resonance (eds. N.R. Krishna and L.J. Berliner), Volume 17, pp. 81–130. Kluwer Academic/Plenum, New York.
- Montelione, G.T., Zheng, D., Huang, Y.J., Gunsalus, K.C., and Szyperski, T. 2000. Protein NMR spectroscopy in structural genomics. Nat. Struct. Biol. 7 982–985. [DOI] [PubMed] [Google Scholar]
- Moseley, H.N.B., Monleon, D., and Montelione, G.T. 2001. Automatic determination of protein backbone resonance assignments from triple resonance nuclear magnetic resonance data. Methods Enzymol. 339 91–108. [DOI] [PubMed] [Google Scholar]
- Murzin, A.G. 1993. OB(oligonucleotide/oligosaccharide binding)-fold: Common structural and functional solution for non-homologous sequences. EMBO J. 12 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri, D., Szyperski, T., Otting, G., Senn, H., and Wütrich, K. 1989. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry 28 7510–7516. [DOI] [PubMed] [Google Scholar]
- Newkirk, K., Feng, W., Jiang, W., Tejero, R., Emerson, S.D., Inouye, M., and Montelione, G.T. 1994. Solution NMR structure of the major cold-shock protein (CspA) from Escherichia coli: Identification of a binding epitope for DNA. Proc. Natl. Acad. Sci. 91 5114–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls, A., Sharp, K.A., and Honig, B. 1991. Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11 281–296. [DOI] [PubMed] [Google Scholar]
- Rost, B. 1996. PHD: Predicting one-dimensional protein structure by profile based neural networks. Methods Enzymol. 266 525–539. [DOI] [PubMed] [Google Scholar]
- Schindelin, H., Jiang, W., Inouye, M., and Heinemann, U. 1994. Crystal structure of CspA, the major cold shock protein of Escherichia coli. Proc. Natl. Acad. Sci. 91 5119–5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schröder, K., Graumann, P., Schnuchel, A., Holak, T.A., and Marahiel, M.A. 1995. Mutational analysis of the putative nucleic-acid binding surface of the cold-shock domain, CspB, revealed an essential role of aromatic and basic residues in binding of single-stranded DNA containing the Y-box motif. Mol. Microbiol. 16 699–708. [DOI] [PubMed] [Google Scholar]
- Stetefeld, J., Jenny, M., Schulthess, T., Landwehr, R., Schumacher, B., Frank, S., Rüegg, M.A., Engel, J., and Kammerer, R.A. 2001. The laminin-binding domain of agrin is structurally related to N-TIMP-1. Nat. Struct. Biol. 8 705–709. [DOI] [PubMed] [Google Scholar]
- Tejero, R., Monleon, D., Celda, B., Powers, R., and Montelione, G.T. 1999. HYPER: A hierarchical algorithm for automatic determination of protein dihedral-angle constraints and stereospecific CβH2 resonance assignments from NMR data. J. Biomol. NMR 15 251–264. [DOI] [PubMed] [Google Scholar]
- Vuister, G.W. and Bax, A. 1993. Quantitative J correlation: A new approach for measuring homonuclear three-bond J(HN-Hα) coupling constants in 15N-enriched proteins. J. Am. Chem. Soc. 115 7772–7777. [Google Scholar]
- Wimberly, B.T., Brodersen, D.E., Clemons Jr., W.M., Morgan-Warren, R.J., Carter, A.P., Vonrhein, C., Hartsch, T., and Ramakrishnan, V. 2000. Structure of the 30S ribosomal subunit. Nature 407 327–339. [DOI] [PubMed] [Google Scholar]
- Wishart, D.S. and Sykes, B.D. 1994. The 13C chemical-shift index: A simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR 4 171–180. [DOI] [PubMed] [Google Scholar]
- Wu, B., Yee, A., Pineda-Lucena, A., Semesi, A., Ramelot, T.A., Cort, J.R., Jung, J.-W., Edwards, A., Lee, W., Kennedy, M.A., et al. 2003. Solution structure of ribosomal protein S28E from Methanobacterium thermoautotrophicum. Protein Sci. (this issue). [DOI] [PMC free article] [PubMed]
- Wunderlich, Z., Acton, T.B., Liu, J., Kornhaber, G., Everett, J., Carter, P., Lan, N., Echols, N., Gerstein, M., Rost, B., et al. 2003. ZebaView: A Web-based tool for organizing the protein target list of the Northeast Structural Genomics Consortium. Protein Sci. (in press) [DOI] [PubMed]
- Yusupov, M.M., Yusupova, G.Zh., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H.D., and Noller, H.F. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292 883–896. [DOI] [PubMed] [Google Scholar]
- Zheng, D., Huang, Y.J., Moseley, H.N.B., Xiao, R., Aramini, J., Swapna, G.V.T., and Montelione, G.T. 2003. Automated protein fold determination using a minimal NMR constraint strategy. Protein Sci. 12 1232–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman, D.E., Kulikowski, C.A., Huang, Y., Feng, W., Tashiro, M., Shimotakahara, S., Chien, C.Y., Powers, R., and Montelione, G.T. 1997. Automated analysis of protein NMR assignments using methods from artificial intelligence. J. Mol. Biol. 269 592–610. [DOI] [PubMed] [Google Scholar]