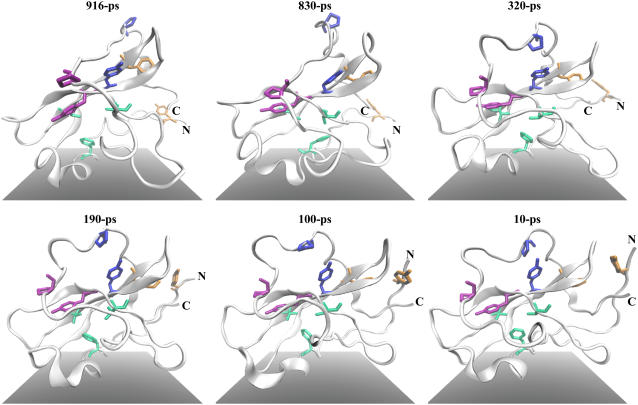
FIGURE 6.
Representative trajectory snapshots taken from the first 500 K thermal unfolding simulation (after root-mean-square fitting on D3 residues), presented in reversed time order to show folding of domain D3. The N- and C-termini are labeled. Folding starts from stabilization of the arch-loop conformation by proline-aromatic interactions between pairs P265-Y245 (magenta sticks) and P271-Y230 (blue sticks) across the loop and the β-folium sheet, denoting event d in Fig. 5. Aromatic-aromatic interaction between F222 and Y190 (on terminal β-sheet, which is also the D2-D3 linker) shown here as orange sticks helps to bind the terminal sheet against the rest of the domain (event c). The tight packing of F202 in the hydrophobic core (F202, V233, and V247 shown as light green sticks) takes the longest time (event b). Note the formation of the terminal β-sheet after F222-Y190 interaction becomes stable. Molecular scenes rendered by Tachyon ray tracer (53) within VMD.