Figure 4. Localization of dynactin subunits in null mutants.
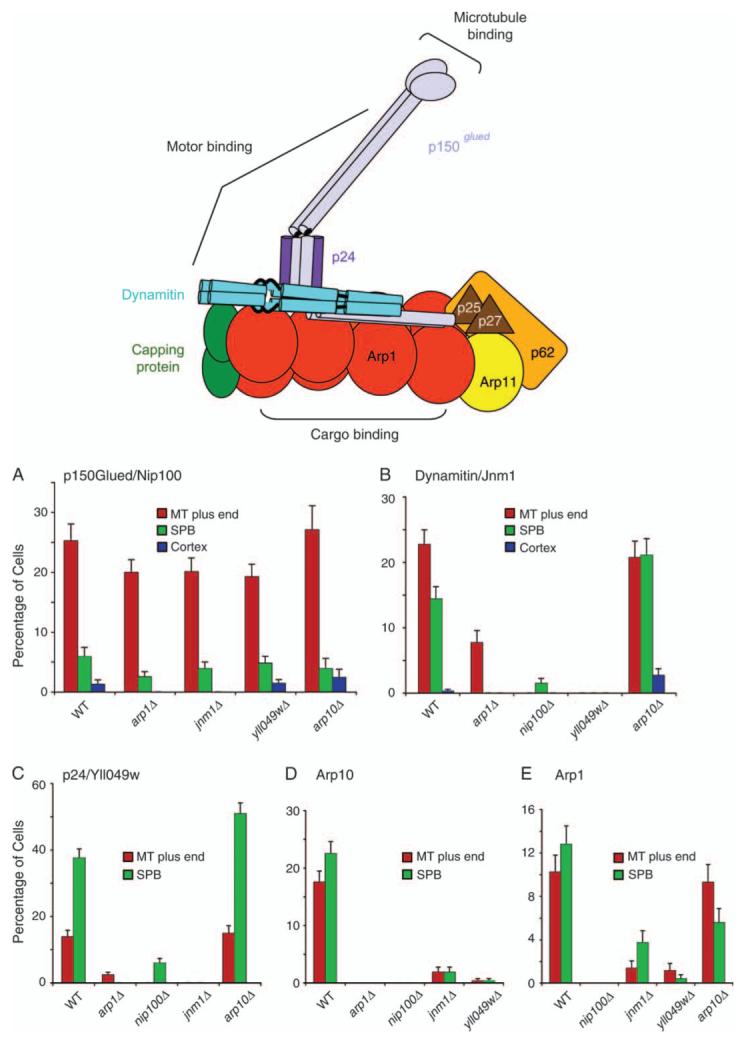
A diagram illustrates the molecular architecture of vertebrate dynactin, modified with permission from one by Trina Schroer (1). On the bar graphs, the percentage of cells that display localization of a fluorescent-tagged subunit to a given location is plotted for different dynactin null mutants. The fluorescent-tagged subunits are A) p150Glued/Nip100-3GFP, B) dynamitin/Jnm1-mCitrine, C) Yll049w-3GFP, D) Arp10-3GFP and E) Arp1-tdimer2. Strain numbers were as follows: A) Wild-type yJC4147; arp1Δ, yJC4158; jnm1Δ, yJC4156; yll049wΔ, yJC4162 and arp10Δ, yJC5377. B) Wild-type yJC5349; arp1Δ, yJC5381; nip100Δ, yJC5385; yll049wΔ, yJC5387 and arp10Δ, yJC5383. C) Wild-type yJC3891; nip100Δ, yJC4166; arp1Δ, yJC4152; jnm1Δ, yJC4154 and arp10Δ, yJC5379. D) Wild-type yJC5389; arp1Δ, yJC5416; jnm1Δ, yJC5418; nip100Δ, yJC5420 and yll049wΔ,yJC5422.E)Wild-typeyJC5400; nip100Δ/nip100Δ, yJC5483; jnm1Δ/jnm1Δ, yJC5482; yll049wΔ/yll049wΔ, yJC5484 and arp10Δ/arp10Δ, yJC5480. MT, microtubule.
