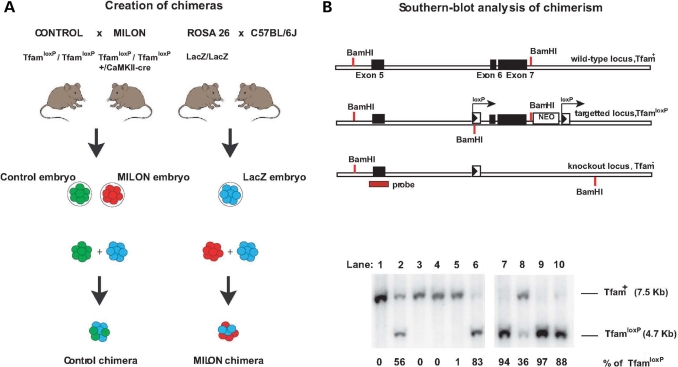
Figure 1.
Creation of mouse chimeras. (A) Control mice were crossed with mitochondrial late-onset neurodegeneration (MILON) mice to generate control embryos (genotype TfamloxP /TfamloxP, indicated by green color) and MILON embryos (genotype TfamloxP/TfamloxP, +/CaMKII-cre, indicated by red color). Homozygous ROSA26 mice (LacZ/LacZ) were crossed with C57BL/6J mice to generate LacZ embryos (genotype +/LacZ, indicated by blue color). Embryo aggregations were performed by mixing control embryos and LacZ embryos or MILON embryos and LacZ embryos to generate control chimeras and MILON chimeras. (B) Genotyping of mouse chimeras using Southern blot analysis. DNA was digested with the restriction endonuclease BamHI and Southern blots were hybridized with a radioactively labeled probe covering the genomic regions surrounding exon 5 of the Tfam gene. The wild-type allele (Tfam+) corresponds to a fragment of 7.5 kb, the non-recombined loxP-flanked allele (TfamloxP) corresponds to a fragment of 4.7 kb and the knock-out allele (Tfam−) corresponds to a fragment of 9.5 kb (upper panel). Genotyping of cerebellar DNA from chimeras was performed to assess levels of chimerism (lower panel). The wild-type Tfam allele (Tfam+) corresponds to cells derived from the LacZ embryo. The loxP-flanked Tfam allele (TfamloxP) corresponds to cells derived from either control embryos or MILON embryos. Additional genotyping to determine the presence of the CaMKII-cre transgene was performed to distinguish between chimeras containing control or MILON cells. The knockout Tfam allele (Tfam−) is not present in cerebellum as cells derived from MILON embryos only express cre-recombinase in neocortex and hippocampus. The ratio between Tfam+ and TfamloxP alleles in cerebellum of individual chimeras thus corresponds to the level of chimerism, expressed as the percentage of TfamloxP alleles.