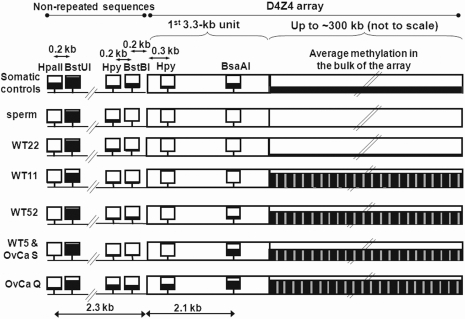
Figure 2.
Cancer-linked hyper- or hypomethylation in the bulk of the D4Z4 array, at the very beginning of the array, and immediately proximal to the array in representative samples. The immediately proximal region includes the 0.8-kb p13E-11 sequence, which is 0.1 kb from the beginning of the D4Z4 array. The squares denote analyzed CpG methylation-sensitive restriction sites. The most proximal HpyCH4IV and BsaAI sites in the first repeat unit of the array are shown followed by a long rectangle representing the foreshortened remainder of the array. The height of the black fill-in is approximately proportional to the average extent of methylation. Methylation in the bulk of the D4Z4 array is diagrammed for HpyCH4IV sites because the small number (six) of HpyCH4IV sites per 3.3-kb repeat unit facilitated quantitation. Other CpG-containing restriction sites were more heavily methylated in somatic controls but gave hypo- or hypermethylation in cancers similar to that seen at HpyCH4IV sites. The gray vertical bars in the D4Z4 array in cancer samples represent the subregion of the D4Z4 repeats units that is partially resistant to hypermethylation and located about 1.4 kb distal to the KpnI site (Figure 3A). See Supplementary Figures 1–3 for most of the data used in this figure.