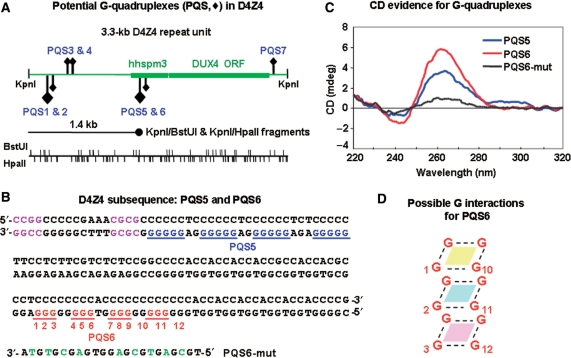
Figure 3.
Potential G-quadruplex sequences (PQS) in D4Z4. (A) Maps of the 3.3-kb D4Z4 repeat unit showing positions of PQS, BstUI and HpaII sites, and the alignment of the unexpectedly strong 1.4-kb band that was observed in BstUI/KpnI and HpaII/KpnI digests (Supplementary Figure 2). Diamonds, PQS; large for G5 and small for G3 runs; upward for those on the forward strand and downward for those on the complementary strand. hhspm3, a sequence that can function as a promoter; DUX4, an open reading frame without a polyadenylation signal (15,16). (B) The D4Z4 subsequence (AF117653) that contains PQS5 and PQS6. Potential unimolecular G-quadruplex structures (blue or red) are shown but there are variations possible in the exact placement of the four runs of G in this subregion that could be involved in unimolecular quadruplexes. In addition, non-adjacent compatible runs of G might participate in quadruplex formation like that seen in bi- and tetramolecular G-quadruplexes. Purple, the closest BstUI and HpaII sites to PQS5; green, mutated residues in PQS6-mut. (C) CD analysis of PQS5, PQS6 and PQS6-mut as single-stranded 23- to 32-nt oligonucleotides. (D) Illustration of one possible orientation for G residues to stack and form a hairpin-type intramolecular G-quadruplex. The numbering corresponds to PQS6 in Panel B, but various other configurations are possible that preserve the planar stacking of four G residues connected by Hoogsteen pairing (dotted lines).