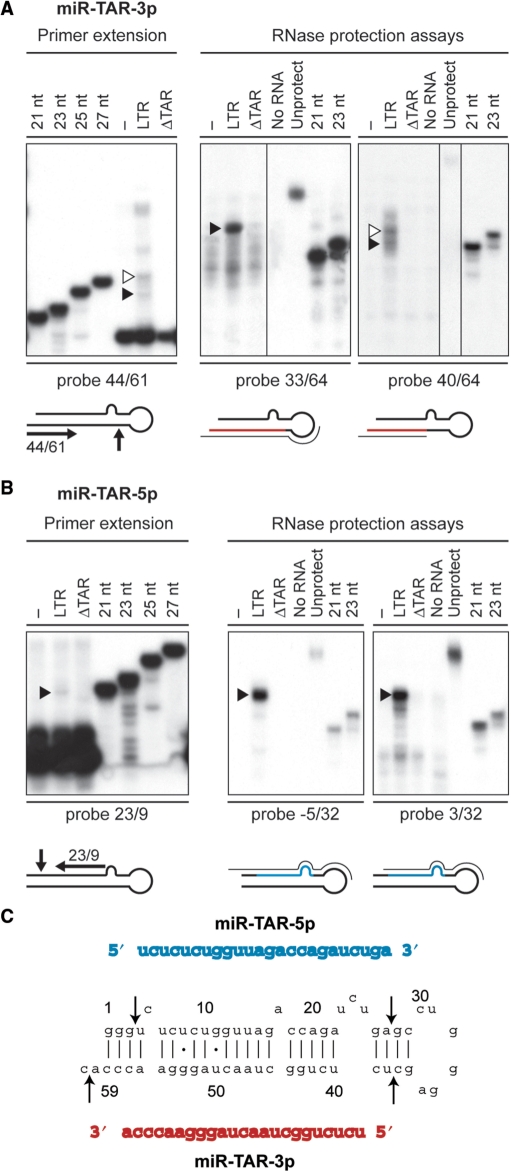
Figure 3.
Delineation of the miRNA duplex derived from the HIV-1 TAR element. (A and B) Small RNAs (<200 nt) isolated from HEK293 cells, transfected with or without pXP2-LTR-Fluc or pXP2-LTRΔTAR-Fluc, were analyzed by primer extension and RPA. (A) miR-TAR-3p. Left, Primer extension analysis using a deoxyribonucleotide complementary to TAR nt 61–44, and run in parallel with DNA size markers. The extended products were analyzed by denaturing PAGE. Right, RPA using RNA probes directed against HIV-1 TAR nt 33/64 and nt 40/64, and run in parallel with RNA size markers. Protected small RNAs were analyzed by denaturing PAGE. Open arrowheads indicate RNA species not supported by concurrent RPA analysis, whereas filled arrowheads indicate the RNA bands considered to establish the consensus TAR miRNA duplex. (B) miR-TAR-5p. Left, Primer extension analysis using a deoxyribonucleotide complementary to nt 23–9. Right, RPA using RNA probes directed against HIV-1 TAR nt −5/32 and nt 3/32. Filled arrowheads indicate the RNA bands considered to establish the consensus TAR miRNA duplex. (C) The consensus miR-TAR-5p:miR-TAR-3p duplex derived from HIV-1 TAR. The arrows indicate the cleavages sites deduced from primer extension and RPA results.