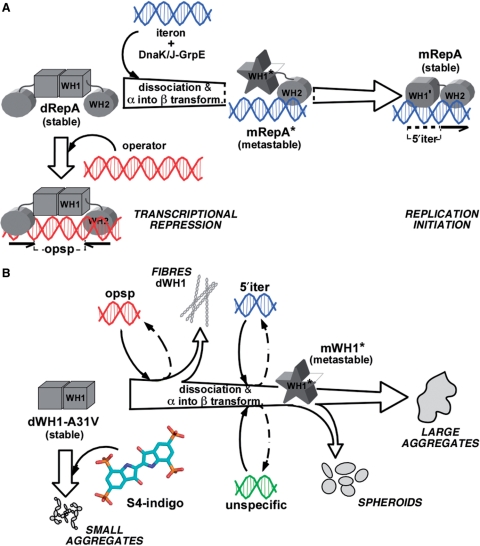
Figure 5.
A scheme of the conformational transactions experienced by pPS10 RepA protein. (A) For the whole RepA protein, the balance between dimers (dRepA) and monomers (mRepA) is modulated by the target iteron DNA sequence (blue). WH2 binding implies dimer dissociation and a local secondary structure conversion (α → β) in WH1 (13,16), which becomes a metastable intermediate (the unbound domain depicted as a star) (14). This short-lived species is stabilized through WH1 binding to the iteron (15). On the contrary, dRepA can bind to the operator DNA sequence (red) without altering its association state and with minor effects (if any) on the structure of the WH1 domain (represented as a cube). Overall, the binding to the iteron sequence defines an irreversible refolding trajectory for WH1. (B) The extent of the remodelling exerted by the core of those target DNA sequences in the WH1 domain, in combination with the amyloidogenic enhancer A31V mutation, lead to the assembly of the protein into either ordered fibres, or spheroids, or large and irregular aggregates (18). Their formation can be interpreted as paralleling the pathway outlined in (A). As it is shown in this article, binding of sulphonated indigoids (S4 molecule depicted) to WH1 dimers competes with dsDNA binding, thus inhibiting amyloidogenesis and leading to small, grainy protein aggregates.