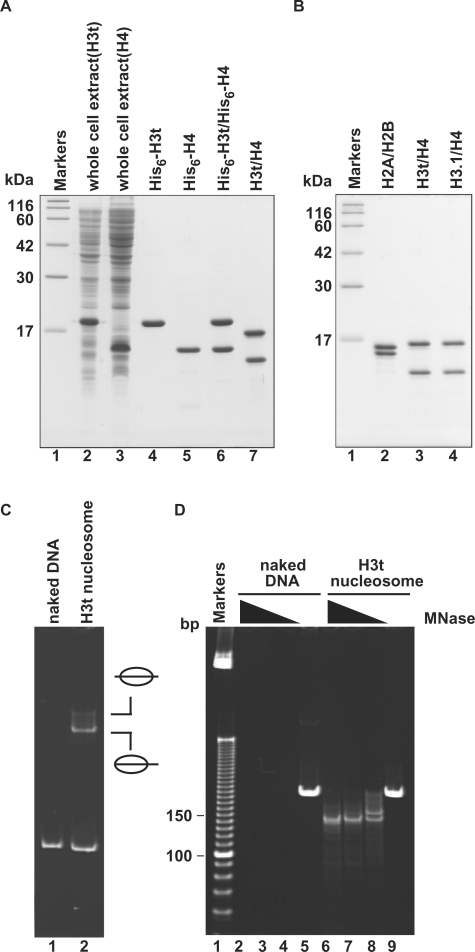
Figure 1.
Preparation and nucleosome formation ability of H3t/H4. (A) SDS–PAGE analysis of proteins from each preparation step for H3t/H4. Lanes 2 and 3: the whole cell lysates of the E. coli cells expressing His6-tagged H3t and His6-tagged H4, respectively. Lanes 4 and 5: the peak Ni-NTA agarose fractions of His6-tagged H3t and His6-tagged H4, respectively. Lanes 6 and 7: His6-tagged H3t/H4 before and after the removal of the hexahistidine tag, respectively. The positions of the size markers (lane1) are indicated on the left. (B) SDS–PAGE analysis of purified histones used in this study. Lane 1: size markers. Lanes 2–4: histones; H2A/H2B (lane 2), H3t/H4 (lane 3) and H3.1/H4 (lane 4). (C) Nucleosome reconstitution analyzed by non-denaturing 6% PAGE. Lane 1: naked DNA. Lane 2: nucleosomes reconstituted with H2A/H2B and H3t/H4 by the salt-dialysis method. Nucleosomes with different translational positions are indicated. (D) MNase assay analyzed by non-denaturing 10% PAGE. Naked DNA (lanes 2–5) and H3t-containing nucleosomes reconstituted by the salt-dialysis method (lanes 6–9) were treated with MNase, and the resulting DNA fragments were analyzed. Lane 1: the molecular mass markers (10 bp DNA ladder). Amounts of MNase (unit/μl) were 0.18 (lanes 2 and 6), 0.13 (lanes 3 and 7), 0.08 (lanes 4 and 8) and 0 (lanes 5 and 9). In H3t-containing nucleosomes, 146 bp DNA, which is tightly wrapped around the nucleosomes and resistant to MNase, is detected (lanes 6–8).