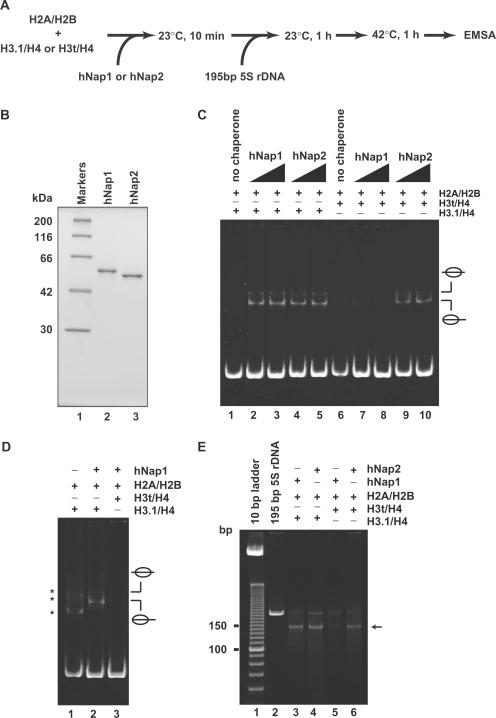
Figure 2.
Nucleosome assembly with H2A/H2B and H3t/H4 or H3.1/H4 by hNap1 and hNap2. (A) Schematic representation of the nucleosome-reconstitution assay by hNap1 or hNap2. (B) SDS–PAGE analysis of purified hNap1 (lane 2) and hNap2 (lane 3) lacking the His6 tag. (C) Nucleosome-reconstitution experiments with the 42°C incubation, analyzed by non-denaturing 6% PAGE. A total of 195 bp 5S DNA was incubated with histones without (lanes 1 and 6) or in combination with hNap1 (lanes 2–3 and 7–8), or hNap2 (lanes 4–5 and 9–10). To assemble nucleosomes containing the histone octamer, H2A/H2B was used with H3.1/H4 (lanes 1–5) or H3t/H4 (lanes 6–10). (D) Nucleosome-reconstitution experiments without the 42°C incubation, analyzed by non-denaturing 6% PAGE. Reactions were performed as in (C). Lane 1 indicates the experiment with H3.1/H4/H2A/H2B in the absence of hNap1. Lanes 2 and 3 indicate the experiments with H3.1/H4/H2A/H2B and H3t/H4/H2A/H2B, respectively, in the presence of hNap1. Bands corresponding to non-specific DNA binding of histones are indicated by asterisks. (E) MNase assay analyzed by non-denaturing 10% PAGE. The samples incubated with the indicated combinations were treated with MNase, and the resulting DNA fragments were analyzed. A total of 146 bp DNA fragments in nucleosomes (arrow) are found in the combinations of H3.1/H4 with hNap1 (lane 3) or hNap2 (lane 4), and H3t/H4 with hNap2 (lane 6), but not H3t/H4 with hNap1 (lane 5).