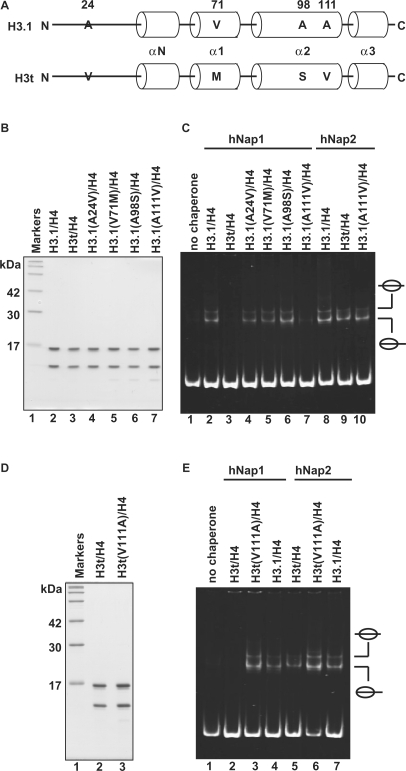
Figure 6.
Mutational analyses of H3.1 and H3t. (A) Sequence comparison between human H3.1 and H3t. The different amino acids between H3.1 and H3t are indicated by capital letters. Cylinders indicate α-helices found in the crystal structure of the human nucleosome core particle (39). (B) SDS–PAGE analysis of the H3.1 mutants complexed with H4. Lane 1: molecular mass markers. Lanes 2 and 3: purified H3.1/H4 and H3t/H4, respectively. Lanes 4–7: H3.1 mutants complexed with H4; lane 4: H3.1(A24V), lane 5: H3.1(V71M), lane 6: H3.1(A98S) and lane 7: H3.1(A111V). (C) Nucleosome-reconstitution with the H3.1 mutants by hNAP1 or hNAP2, analyzed by non-denaturing 6% PAGE. A total of 195 bp 5S DNA was incubated with hNap1 (lanes 2–7) or hNap2 (lanes 8–10) in combination with H3.1/H4 (lanes 2 and 8), H3t/H4 (lanes 3 and 9) and H3.1 mutants/H4 (lanes 4–7 and 10); all reactions were performed in the presence of H2A/H2B. (D) SDS–PAGE analysis of the H3t(V111A) mutant complexed with H4. (E) Nucleosome-reconstitution with H3t(V111A)/H4 by hNap1 or hNap2, analyzed by non-denaturing 6% PAGE. A total of 195 bp 5S DNA was incubated with hNap1 (lanes 2–4) or hNap2 (lanes 5–7) in combination with H3t/H4 (lanes 2 and 5), H3t(V111A)/H4 (lanes 3 and 6) and H3.1/H4 (lanes 4 and 7); all reactions were performed in the presence of H2A/H2B.