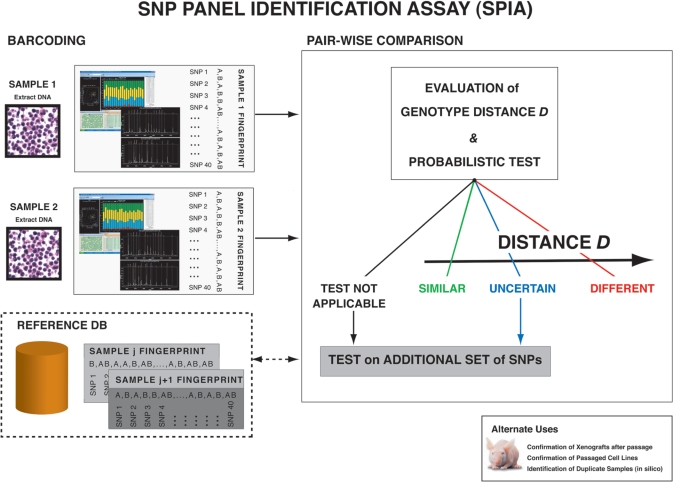
Figure 2.
Schema of SNP panel identification assay (SPIA) applicability and use modality. One typical scenario would be to check out of the identity of a cell lines (SAMPLE 1) with respect to already fingerprinted cell lines, the data of which are stored in the BARCODING REFERENCE database. Another scenario would be to determine the identity of two cell lines, for example derived from the same patients, one from benign tissue cells and one from cancer tissue cells (SAMPLE 1 and SAMPLE 2). First, the DNA is extracted from the cell line(s) and is experimentally genotyped along the set of specified loci included in the SNP panel. Sequenom mass spectrometric technology, allele-specific PCR or other techniques can be implemented. Afterwards, the allele call output sets (DNA fingerprints) are compared. Comparison is performed pair-wise. Comparison with all the fingerprints stored in the reference database can be performed on demand. The SPIA test provides the genotype distance between the two samples and detailed information on the allele mismatches. It also provides the user with a probability measure of the test output, scoring the tested pair as ‘similar’ or ‘different’. If the test result is uncertain, the assay can be repeated using an additional SNP panel.