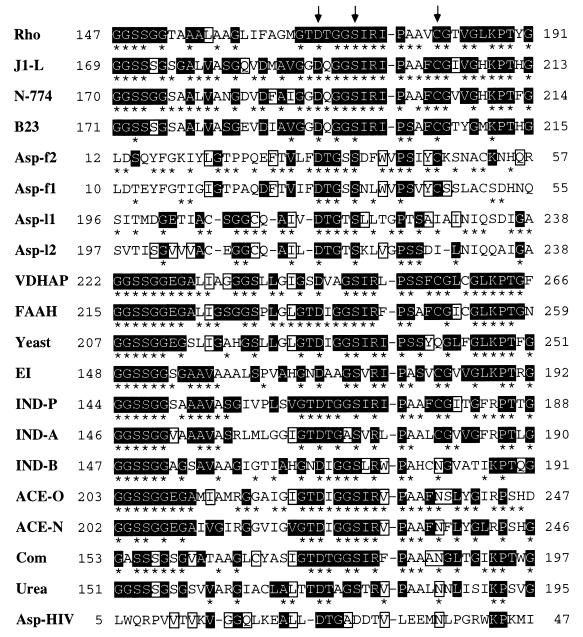
Figure 3.
Alignment over the amidase signature sequence region (3, 13, 17) of all members of the amidase family previously reported with the aspartic proteinases. From top to bottom (numbers of the total amino acids of each overall sequence and their accession numbers of databases are given in brackets just after each enzyme name): Rho, amidase from Rhodococcus sp. [462 residues, GenBank M74531 (17)]; J1-L, amidase from R. rhodochrous J1 [515 residues, GenBank D16207 (10)]; N-774, amidase from Rhodococcus sp. N-774 (whose sequence is identical with that of the Brevibacterium R312 amidase) [521 residues, GenBank X54074 or M60264 (19, 20)]; B23, amidase from Pseudomonas chlororaphis B23 [506 residues, GenBank D90216 (21)]; Asp-f2, the N-terminal region of calf chymosin [323 residues, PIR CMBO (43, 44)]; Asp-f1, the N-terminal region of porcine pepsin [325 residues, GenBank J04601 (45)]; Asp-l1, the C-terminal region of porcine pepsin [325 residues, GenBank J04601 (45)]; Asp-l2, the C-terminal region of calf chymosin [323 residues, PIR CMBO (43, 44)]; VDHAP, a vitamin D3 hydroxylase-associated protein from cockerel [464 residues, GenBank U00694 (13)]; FAAH, oleamide hydrolase from rat [579 residues, GenBank U72497 (3)]; Yeast, putative amidase from Saccharomyces cerevisiae [549 residues, GenBank X56043 (46) (only this putative amidase is described here as a representative of putative amidases; two other putative sequences of Caenorhabditis elegans are not shown here)]; EI, EI enzyme from Flavobacterium [493 residues, GenBank M26953 (15)]; IND-P, indoleacetamide hydrolase from P. savastanoi [455 residues, GenBank M11035 (4)]; IND-A, indoleacetamide hydrolase from Agrobacterium tumefaciens [467 residues, PIR Q2AGAT (5)]; IND-B, indoleacetamide hydrolase from Bradyrhizobium japonicum [465 residues, GenBank X15117 (47)]; ACE-O, acetamidase from Aspergillus oryzae [545 residues, GenBank D10492 (48)]; ACE-N, acetamidase from Aspergillus nidulans [548 residues, GenBank M16371 (49)]; Com, amidase from Comamonas acidovorans KPO-2771–4 [473 residues (8)]; Urea, urea amidolyase from Candida utilis [1830 residues, PRF 2009329A (34)]; Asp–HIV, aspartic proteinase from HIV type-1 [102 residues, GenBank HIVU53613 (50)]. Arrows indicate the amino acid residues that correspond to Asp191, Ser195, and Cys203 of the R. rhodochrous J1 amidase examined in this experiment. Numbers on the left and right indicate the position of the amino acid residue in each molecule. Gaps (-) were inserted to increase sequence similarity. Identical residues are denoted by asterisks between neighboring sequences. Residues highlighted in reverse type are conserved across both amidases and aspartic proteinases in at least four of 20 sequences, and the first four residues GGSS highlighted in reverse type in most of the amidases do not appear in the aspartic proteinases with the exception of Ser14 in Asp-f2. Except the residues highlighted in reverse type, identical amino acid residues, all of which are not conserved between both aspartic proteinase and amidase families, are boxed.