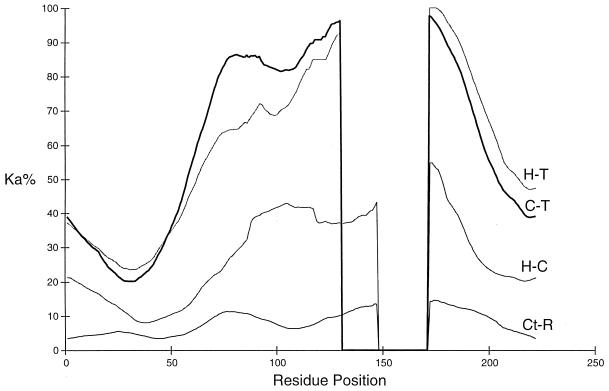
Figure 4.
Plot of nonsynonymous substitution percent (Ka%) against amino acid residue position (numbering follows that of Fig. 2) in selected pairwise comparisons of human (H), cattle (Ct), rat (R), caiman (C), and toad-2 (T) amelogenin sequences. An initial sliding window of 30 codons was used to estimate Ka by the method of Li et al. (32), and a further sliding window of 30 codons was used to smooth the values obtained. Small alignment gaps (<8 residues) were ignored in the plot; larger gaps are indicated by vertical lines.