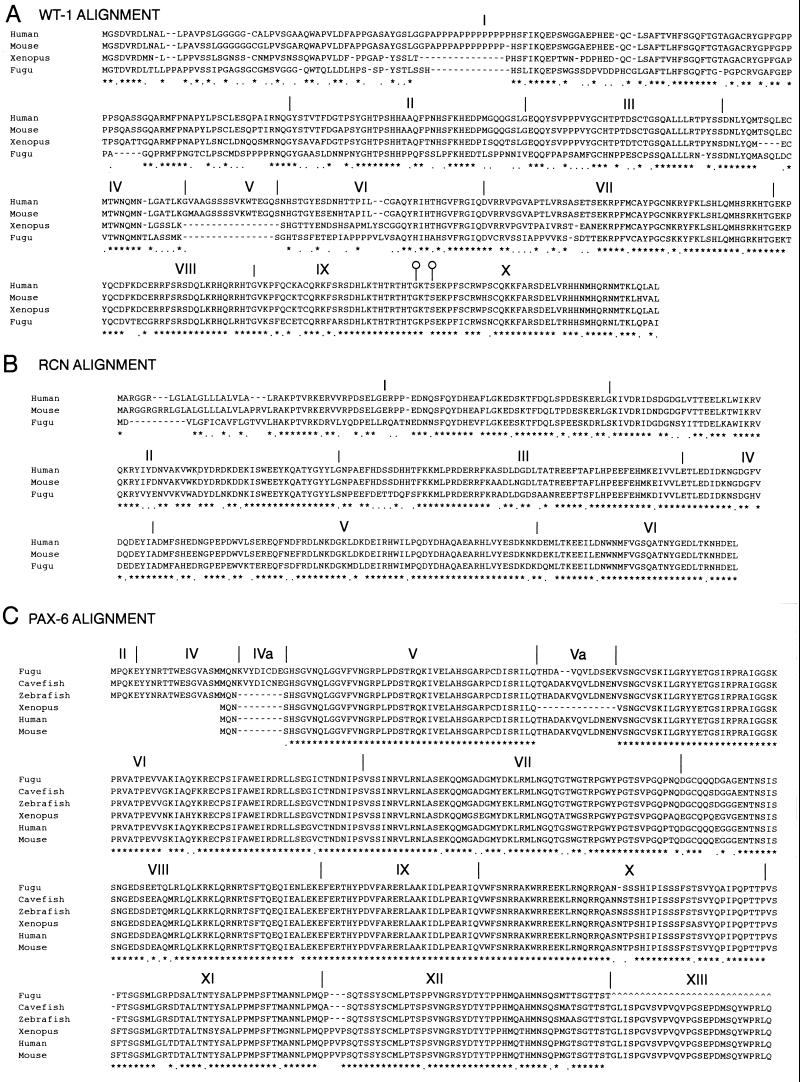
Figure 2.
(A) Clustal alignment of WT1 genes from human, mouse, Xenopus, and Fugu. Exons are numbered and their boundaries marked by a vertical line in all three alignments. Both Xenopus and Fugu genes lack the polyproline tract in the middle of exon 1 and the whole of the alternatively spliced exon V. The alternative splice donor sites at the end of exon IX are marked by open circles on the boundary markers. (B) Clustal alignment of mouse, human, and Fugu RCN1 genes. Intron/exon boundaries are marked for Fugu as these are not published for the mouse or human genes. It is likely that the mammalian genes have a very similar or identical genomic organization. (C) Clustal alignment of PAX6 genes from Fugu, blind cavefish, zebrafish, Xenopus, mouse, and human. In the three fish species shown, there are additional coding sequences at the 5′ end of the gene giving rise to a variety of transcripts. The most 5′ transcripts in fish are formed by splicing together exons II and IV, skipping exon III. There is also an additional exon found in cavefish and (by homology) Fugu that has not been found in higher vertebrates and does not appear to be present in human genomic sequence for this region (HSCFAT5 and HSA1280). Exon 13 has not yet been sequenced in Fugu.