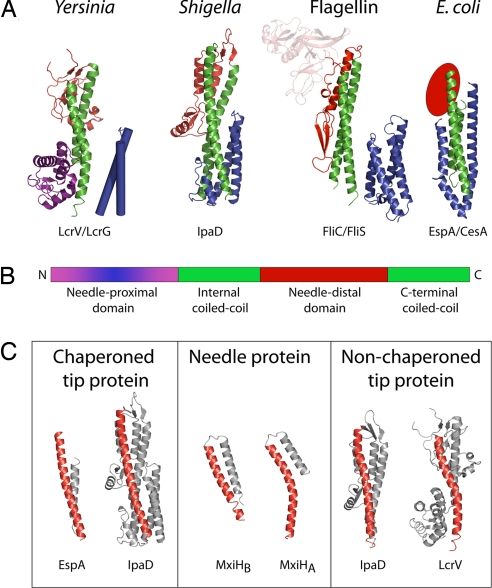
Fig. 2.
Needle tip-associated adaptor protein structures. (A) Cartoon representation of the four classes of adaptor proteins thus solved: Yersinia family (LcrV; PDB ID code 1R6F) (41); Shigella family (IpaD; PDB ID code 2J0O) (39); Flagellin family (FliC; PDB ID code 1IO1) (22); E. coli family (EspA; PDB ID code 1XOU) (42). The structures demonstrate a common architecture, consisting of a central coiled-coil (green), a needle-distal domain (red), and a needle-proximal domain (purple/blue). The needle-proximal portion of the coiled-coil, which is postulated to be involved in assembly at the needle-tip, is chaperoned (shown in blue). This chaperoning may be via an intramolecular mechanism (IpaD: IpaD N-terminal domain) or separate chaperones [EspA:CesA, PDB ID code 1XOU (42), FliC:FliS, PDB ID codes 2IO1/1ORJ (22, 40), LcrV:LcrG, LcrV; PDB ID code 1R6F; no structure available for LcrG, modeled on IpaD N-terminal domain structure]. (B) Schematic representation of the common architecture. (C) The C-terminal helix of the central coiled-coil is straight in chaperoned adaptor proteins (IpaD chaperoned, PDB ID zj0o and bent in the nonchaperoned structure (IpaD nonchaperoned, PBD ID 2j0n). This bend is comparable to the kink in the C-terminal helix of the Shigella needle protein MxiH (PDB ID code 2CA5) (24).