Fig. 2.
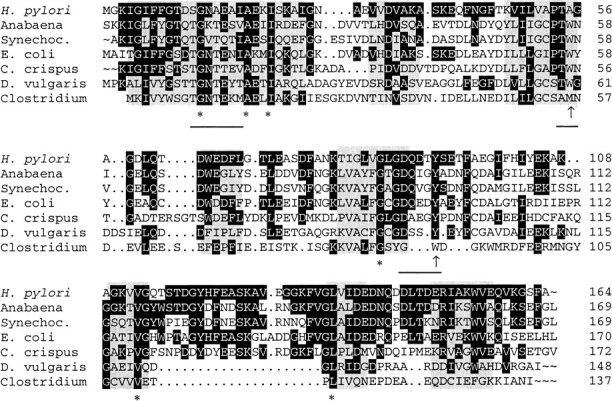
Structural alignment of flavodoxins with resolved three-dimensional structure. The alignments were performed according to regions of structural similarity as determined by Multiple Alignment of Proteins Structures (MAPS) (Guoguang 1998). Structural identity between all crystal structures is indicated by gray boxes. The 6 amino-acid residues conserved between these proteins are marked by asterisks, while amino acids identical to the Helicobacter pylori flavodoxin are shown with a black background. Hydrophobic amino acids in proximity to the isoalloxazine ring are indicated by an arrow, and residues involved in binding of the FMN cofactor are underlined.
