Abstract
Streptococcal protein G (SpG) is a bacterial cell surface receptor exhibiting affinity to both human immunoglobulin (IgG) and human serum albumin (HSA). Interestingly, the serum albumin and immunoglobulin-binding activities have been shown to reside at functionally and structurally separated receptor domains. The binding domain of the HSA-binding part has been shown to be a 46-residue triple α-helical structure, but the binding site to HSA has not yet been determined. Here, we have investigated the precise binding region of this bacterial receptor by protein engineering applying an alanine-scanning procedure followed by binding studies by surface plasmon resonance (SPR). The secondary structure as well as the HSA binding of the resulting albumin-binding domain (ABD) variants were analyzed using circular dichroism (CD) and affinity blotting. The analysis shows that the HSA binding involves residues mainly in the second α-helix.
Keywords: Binding, affinity, human serum albumin (HSA), albumin-binding domain (ABD), mutational analysis
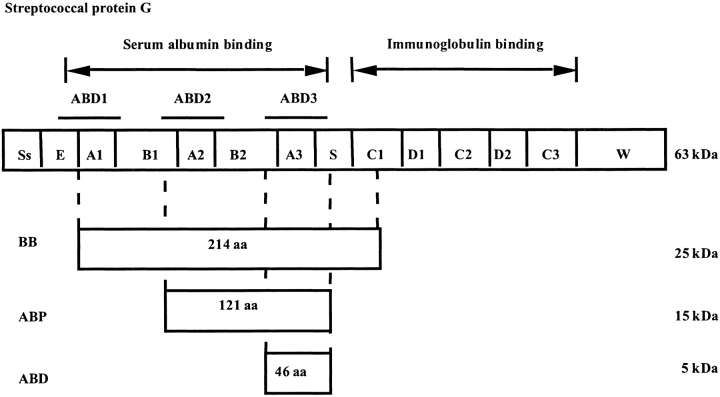
Several species of both Staphylococcus and Streptococcus have evolved receptors with affinity to various proteins, such as immunoglobulins (Forsgren and Sjöquist 1966), albumin (Kronvall et al. 1979), and plasmin (Lottenberg et al. 1987). Streptococcal protein G (SpG) is a bacterial receptor displayed on the surface of Streptococcus strain G148. The extracellular part of SpG consists of one immunoglobulin (IgG)-binding region and one serum albumin-binding region (Fig. 1 ▶). These different regions have three independently folding domains with affinity to either albumin or immunoglobulin (Olsson et al. 1987). The IgG-binding part has been widely used for affinity purification of immunoglobulins.
Fig. 1.
An overview of the streptococcal protein G (SpG). The IgG and albumin-binding parts are separated. Both parts consist of three homologous domains. Nomenclature according to Ståhl and Nygren (1999).
The albumin-binding part is also interesting for several reasons. The domain can be used as affinity ligand for purification of human serum albumin (HSA) and also as a fusion partner for affinity purification of other target molecules (Hammarberg et al. 1989). Fusing the albumin binding part to several vaccine candidates has shown to increase the stability in serum (Nygren et al. 1991; Makrides et al. 1996). In addition, an increased immunogenicity has been reported for the respiratory syncytial virus (RSV) vaccine candidate BBG2N, when fusing the vaccine moiety to the albumin-binding domain (ABD) (Sjölander et al. 1997; Libon et al. 1999).
The three homologous HSA-binding domains have similar sequences and are designated ABD1, ABD2, and ABD3 (Fig 1 ▶). The structure of one of the ABD domains (ABD3) has been determined by NMR spectroscopy to be a left-handed three-helix bundle. It consists of 46 amino acids and is stable enough to be independent from disulfide bridges, bound ligands, cross-links, or metal ions (Kraulis et al. 1996). The ABD domain is chemically and thermally stable and therefore, is a suitable scaffold for further engineering. The resistance toward alkaline conditions has also been improved by replacing the aspargine residues by protein engineering (Gülich et al. 2000).
Despite its biotechnological importance, the interaction between ABD and HSA has not yet been elucidated. Interestingly, the three-dimensional structures of both interacting components are known. The structure of HSA was determined by X-ray crystallography (Carter and He. 1990; Curry et al. 1998), whereas the structure of ABD was determined by NMR (Kraulis et al. 1996). To understand the molecular basis of this protein–protein recognition, a detailed characterization of individual interactions in the interface between the proteins is required. Therefore, we have characterized the molecular basis for the interaction by protein engineering of the ABD domain using an alanine scanning procedure. Alanine scanning is a well-established method and has been widely used to determine the binding properties for different molecular interaction partners (Bass et al. 1991; Cunningham and Wells 1993; Jendeberg et al. 1995; Schreiber and Fersht 1995; Gårdsvoll et al. 1999).
In this work, we evaluated the affinity and the kinetics by the use of surface plasmon resonance (SPR). This method measures the binding of the ligand to an immobilized target. It provides a real-time measurement of the events in the binding and releasing procedure. The SPR-technique has been used to determine the relative effect of different mutations on the kinetic constants. The results suggest that the binding site of ABD toward HSA is located mainly on the second helix.
Results
General strategy
To analyze which part of ABD that binds to HSA, an alanine-scanning procedure was performed in a consecutive manner. Initially, residues in position E3, Y20, E32, and E40 were replaced. These amino acids were chosen as they were surface exposed, situated on all three α-helices, and also pointing in different directions. To determine more precisely what amino acids were involved in the interaction, further alanine substitutions around the most participating residues were made.
Expression and purification of ABD variants
To simplify the purification procedure the ABD-variants were expressed as fusions to the IgG-binding domain Z, based on the staphylococcal protein A domain (Nilsson et al. 1987). All Z-ABD variants were successfully produced as intracellular proteins in Escherichia coli at 37°C and show the same expression levels, ∼50 mg/L as estimated from SDS-PAGE. Z-ABD and the mutants thereof could efficiently be purified on IgG columns. This indicates that the fused domains folded correctly and were not influenced by their fusion partners to any great extent. Depending on the ability of the mutant to bind HSA, the second purification was either on an HSA affinity column or in a reversed-phase chromatography (RPC) procedure. After the second purification, samples were analyzed with SDS-PAGE, lyophilized, and stored for further analyses.
Structural characterization of different ABD variants
Structural analyses were performed on a circular dichroism (CD) equipment, as it previously has been proven to be suitable for detecting structural changes in α-helical proteins (Johnson 1990; Nord et al. 1997). Purified fusion proteins were subjected to a subtractive circular dichroism spectroscopy analysis in which the signal contribution from the Z domain was subtracted (Nord et al. 1997). All spectra show a minimum at 208 nm and a shoulder at 222 nm in combination with a maximum around 195 nm, indicating high α-helical content in the proteins. The results from the CD measurements show that the backbone configuration is similar for all the different ABD variants indicating the structure to be preserved (data not shown).
Biospecific interaction analysis
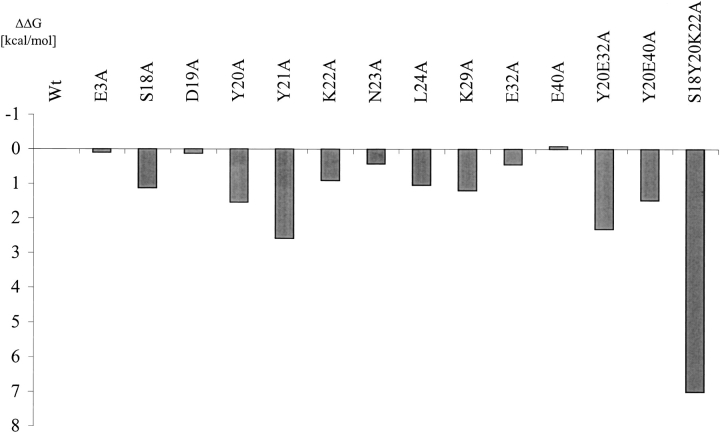
To determine the differences in affinity for the ABD variants toward HSA, SPR, using a Biacore, was carried out. The aim was to compare the affinity for the different mutated ABD variants with the parental molecule. By using the same concentration for all proteins we were able to analyze how the affinity changed for the different mutations introduced. Of the first four mutated amino acids, Y20 was shown to have most influence on the binding to HSA (Table 1 and Fig. 2 ▶).
Table 1.
An overview of the kinetic study of the ABD variants carried out on the Biacore
| ZABD variant | koff [10−3 s−1] | kon [105 M−1 s−1] | Kaff [107 M−1] | δδG [kcal/mol] |
| wt | 0.6 (0.2) | 1.4 (0.2) | 25.9 | 0.0 (0.2) |
| E3A | 0.6 (0.3) | 1.4 (0.7) | 22.2 | 0.1 (0.1) |
| S18A | 3.5 (1.6) | 1.3 (0.3) | 3.9 | 1.1 (0.1) |
| D19A | 0.8 (0.3) | 1.5 (0.3) | 20.9 | 0.1 (0.3) |
| Y20A | 5.1 (2.1) | 0.9 (0.2) | 1.9 | 1.5 (0.2) |
| Y21A | 3.9 (2.4) | 0.1 (0.07) | 0.3 | 2.6 (0.2) |
| K22A | 1.3 (0.7) | 0.7 (0.2) | 5.6 | 0.9 (0.2) |
| N23A | 0.9 (0.3) | 1.0 (0.1) | 12.7 | 0.4 (0.1) |
| L24A | 0.6 (0.4) | 0.3 (0.02) | 4.5 | 1.0 (0.2) |
| K29A | 1.1 (0.4) | 0.3 (0.1) | 3.4 | 1.2 (0.6) |
| E32A | 2.4 (0.9) | 2.5 (0.3) | 12.2 | 0.4 (0.3) |
| E40A | 1.2 (0.5) | 2.6 (1.5) | 29.5 | −0.1 (0.6) |
| Y20E32A | 17 (11) | 0.5 (0.2) | 0.5 | 2.3 (0.1) |
| Y20E40A | 6.8 (2.1) | 1.4 (0.1) | 2.1 | 1.5 (0.1) |
| S18Y20K22A | 6 | 0.0001 | 0.0002 | 7 |
Standard deviations are given in parentheses. Since the very low affinity of the triple mutant leads to problems in the detection of the binding, no standard deviation is given for that specific mutant.
Fig. 2.
Graphic comparison of the changes in free binding energy of the analyzed ABD variants compared to the parental ABD (δδG in kcal/mole). The values are calculated using BIAevaluation 3.02.
Because residues 18–24 are close to tyrosine 20, they were chosen for further analysis. Also, lysine 29 is surface exposed and in the vicinity of tyrosine 20, and therefore, could take part in the HSA binding. Hence, the following single mutants were made: S18A, D19A, Y21A, K22A, N23A, L24A, and K29A. To further confirm the variations between all the mutated ABD variants, the binding patterns for all these mutants were analyzed using SPR.
The binding analysis shows that 8 of 11 single mutants were significantly affected in binding (S18, Y20, Y21, K22, N23, L24, K29, and E32) (Table 1). From these data, the differences in free binding energy were calculated (Fig. 2 ▶). Mutations on position S18, Y20, Y21, K22, N23, L24, K29, and E32; all decrease the affinity. Of these, Y20 and Y21 were shown to have most influence on the affinity toward HSA (Table 1 and Fig. 2 ▶). These residues are both located in the amino-terminal part of the second helix and are also surface exposed. There is, however, one single mutant, E32A, which has both a higher on-rate and off-rate, compared to the parental ABD. However, the contribution of these two parameters does not significantly affect the total binding energy. A mutation can have a specific effect on kon or koff, respectively (Schreiber and Fersht 1995). This behavior has also been observed previously in the analysis of the binding site of human growth hormone (hGH) receptor (Cunningham and Wells 1993).
In the case of D19A, we were not able to detect any significant change either in on-rate or off-rate (Table 1). Hence, no important interactions between the aspartic acid and HSA are formed in the complex. A substitution of amino acid K29, which is in the vicinity of Y20 and Y21, has an evident effect on the on-rate. This is probably because the positive charge is removed and thereby the attraction is decreased between the proteins when docking. This behavior can be explained by the fact that both HSA and ABD are negatively charged at the pH used in the binding studies. Taking a positive charge away will, therefore, lead to a slower docking of the complex. A similar effect could be detected when mutating E32 for an alanine. A small increase in the on-rate was measured, probably due to removal of a negative charge, which decreases the repulsion between HSA and ABD. Also, a rather large increase in off-rate was detected, indicating that this amino acid interacts with HSA when the complex is formed. This behavior has also been discussed in a paper by Albeck and Schreiber (1999).
To analyze the effect of multiple mutations on structure and affinity, three additional mutants were made: ABD (Y20A, E40A), ABD (Y20A, E32A), and ABD (S18A, Y20A, K22A). By comparing the double mutants with the corresponding single mutants we were able to conclude that the effects for the changes in affinity for residues Y20, E32, and E40 were rather additive, indicating that neither the single nor the double mutations affect the surrounding amino acids to any great extent. By comparing the change in affinity for the corresponding single mutants (S18, Y20, and K22) with the triple mutant ABD (S18A, Y20A, K22A) one can observe a synergistic effect, indicating that in the triple mutant the introduced mutations also have affected the residues located close to the mutations. However, most probably, the same substitutions made in single mutants do not greatly affect other residues in the close vicinity.
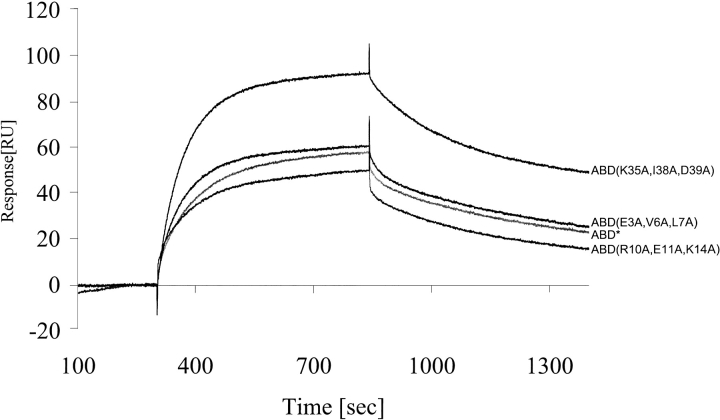
To further verify the position of the binding surface, we made three mutants where three neighboring amino acids were changed at a time. Mutations for alanine were made in helix one and three, in positions where the amino acids were pointing away from the postulated binding site. The resulting proteins were: ABD* (E3A, V6A, L7A), ABD* (R10A, E11A, K14A), and ABD* (K35A, I38A, D39A). All three mutants were shown to have similar or better affinity to HSA than the parental molecule (Fig. 3 ▶).
Fig. 3.
The binding pattern for the three triple mutants of ABD*, E3A, V6A, L7A; R10A, E11A, K14A; and K35A, I38A, D39A. The concentration was 100 nM. The signal from a nonimmobilized surface was subtracted. The mean values from at least two different measurements are shown.
Discussion
A protein engineering approach was used to characterize the protein–protein interaction between ABD and HSA. The different ABD variants were produced in high yield and efficiently purified using the IgG-binding domain Z (Nilsson et al. 1987). All together 14 new variants of ABD were cloned and expressed. Their structure and affinity toward HSA were analyzed. Of these 14 variants, 11 of the constructs showed decreased affinity and 3 were not significantly affected.
The ABD variants have shown similar secondary structure according to the CD measurements, both regarding the position of the minima and the amplitude of the signals. The differences in the amplitude between the spectra are within the expected error of the determination of the protein concentration. However, because the exact structures of the different mutants are unknown, minor changes of structure due to the mutations cannot be ruled out (Clackson et al. 1998; Vaughan et al. 1999).
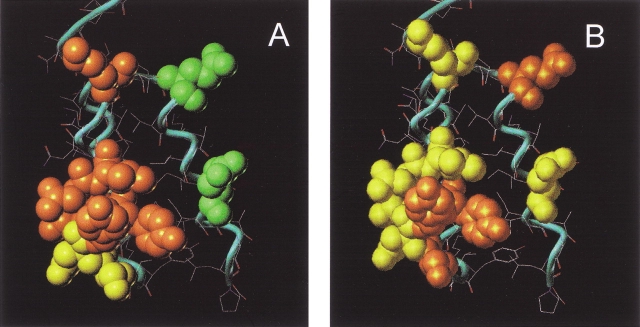
By comparing the change in association and dissociation rates for the analyzed ABD variants, it is possible to elucidate the participation of the different amino acids in the binding mechanism. Different amino acids contribute differently to the on-rate and off-rate (Table 1). When looking at the different on-rates, almost all mutations made in the second helix have a negative contribution (Fig. 4A ▶). These results suggest that major parts of the second helix are directly or indirectly involved in the first step of the binding mechanism between ABD and HSA (Otzen and Fersht 1999; Albeck et al. 2000). The changes in the rate of the docking of the complex could be explained either by a structural perturbation or an electrostatic change of ABD due to the mutations. However, it is reasonable that some of the changes in the on-rate can be explained by electrostatic influences. Because both interacting molecules are negatively charged during the analyses, the association kinetics are highly sensitive to changes in the surface charge. This has also been shown by Albeck and Schreiber (1999). An electrostatic change of the molecule could also be the explanation why kon of ABD* decreases, whereas koff is unaffected (Gülich et al. 2000). In ABD*, four slightly positively charged aspargines are replaced by one leucine, two aspartic acids, and one lysine. Because the small decrease in affinity mainly is due to the lower on-rate, it is probably the change in charge of the entire molecule that is responsible (Gülich et al. 2000). In the third helix, a substitution on both position 32 and 40 results in a slightly increased kon. Although the change in on-rate for the E40A mutant not is strictly statistically reliable we believe that this conclusion is further supported by the on-rate measured for Y20A, E40A. For this mutant we are not able to detect the decrease in on-rate that would be expected from the Y20A mutation, probably due to a compensating effect from the E40A mutation (Table 1 and Fig. 4A ▶). A plausible explanation is a long-range charge repulsion. Replacing a negative charge with an alanine would decrease the repulsion and increase the kon. The dissociation kinetics, however, are not explained by any simple rules. This is probably because the interactions in the binding surface are of a highly cooperative nature and also very specific.
Fig. 4.
The three-dimensional structure of ABD where the effect of different mutations is displayed. (A) Change in kon upon mutation. Orange color means a decreased and green color shows an increased on-rate when the original residue is exchanged to alanine. Residues shown in yellow denote an unchanged behavior in the on-rate despite a change to alanine. (B) How the off-rate changes while replacing certain residues for alanine. Orange shows a position where the off-rate increases and green color positions where the off-rate decreases upon mutation. Yellow means positions where a mutation does not affect the koff.
Three additional ABD variants were made to further define the surface that interacts with HSA. All three proteins were made as triple mutants with ABD* as the parental molecule. However, by comparing the binding kinetics of the triple mutants with the parental ABD* (Gülich et al. 2000), we were able to assess the importance of these nine amino acids in the association and dissociation process. The binding analyses show that the off-rate is almost identical for all three mutants, indicating that the stability of the ABD–HSA complex has not been affected. However, one of the mutants shows an increased on-rate (Fig. 3 ▶). This can be explained by the change from a negatively charged aspartic acid to an uncharged alanine. In this triple mutant, a positively charged lysine was also changed for an alanine. The positive effect on the overall binding confirms that this lysine is situated away from the binding, thus not affecting the affinity to HSA.
Hence, possibly a small part of the ABD domain is responsible for the binding to HSA. Residues Y20 and Y21 have the largest effect on the affinity (see Table 1 and Fig. 2 ▶) and by changing only three amino acids (S18, Y20, and K22), we were able to remarkably diminish the binding site (Table 1 and Fig. 2 ▶). In fact, the triple mutant ABD (S18A, Y20A, and K22A) has lost 105 in affinity compared to wild type (wt) (Table 1). The effect of this triple mutant in the affinity to HSA is larger than expected from the corresponding single mutants. This behavior could possibly be due to coordinating effects in orientation of the directly interacting amino acids (Clackson et al. 1998; Vaughan et al. 1999). To elucidate the exact contribution of each amino acid a structural determination of the interacting molecules is essential. However, the large lost in affinity for this triple mutant strongly supports the theory that most of the interaction between ABD and HSA is located in the second helix. This is in contrast to earlier studies by Johansson et al. (1997), which propose that these positions could not be involved in the binding to HSA. In a hydrogen–deuterium exchange experiment, residues 18, 19, 20, 31, and 32 exhibited fast exchange in the complex, protein PAB and HSA. Protein PAB is a molecule with similar structure and function as ABD. Our results show that at least three of these positions are involved in the stabilization of the complex. We can also conclude that helix 1 and also most parts of helix 3 are probably not involved in the binding of HSA.
Here, we have located the binding site of ABD toward HSA. These results give us new ways to enhance and improve ABD for different applications. A few amino acids were shown to be responsible for most of the affinity of ABD and most amino acids that affected the affinity in a major way were located in the second helix. Hereby we are able to introduce new binding specificity in the small domain, but still retain the affinity for HSA.
Materials and methods
Cloning
A plasmid encoding ABD, pTrpABD (Kraulis et al. 1996) was used as cloning vector. To simplify the purification of ABD, a gene encoding the Z domain from protein A (Nilsson et al. 1987) was introduced at the amino-terminal of ABD. The Z domain was taken from pRIT45 by using restriction enzymes XbaI and EcoRI and ligated into pTrpABD restricted with the same enzymes. The new vector was entitled pTrpZABD.
To introduce the different mutations in ABD, a two-step PCR technique was used, with pTrpZABD as template (Higuchi et al. 1988). All oligonucletiodes used were purchased from Interactiva, Germany. To confirm the defined location of the binding site, three triple mutants were also constructed and produced. This was done by introducing the mutations in the vector pTrpABD* (Gülich et al. 2000). In the first triple mutant the mutations were made at position E3, V6, and L7, in the second R10, E11, and K14, and in the third K35, I38, and D39. All mutations introduced alanine instead of the wild-type amino acid. To ensure that the DNA sequence was correct, a DNA-sequencing procedure was carried out, using the BigDye terminator method on the ABI 377 platform (Perkin Elmer Applied Biosystems Division, Foster City, CA). The 17 variants were named according to the substitutions.
Production and purification
The cultivation and production was performed as described by Kraulis et al. (1996). The purification of the fusion proteins was done by affinity chromatography on IgG columns as described by Nilsson et al. (1987). To further purify the proteins, an additional purification step was applied using HSA affinity chromatography of the mutants that still retained the affinity for HSA (Nygren et al. 1988). The HSA-affinity for the different mutants was analyzed by an affinity blotting technique using labeled HSA (see below). For mutants that have lost most of the affinity toward HSA, the second purification step was carried out by reverse-phase high performance liquid chromatography (RP-HPLC). Samples were loaded on a C18 column, (Vydac), at a flow rate of 1 mL/min at 40°C. The column was previously equilibrated with 33% acetonitril supplemented with 0.25% pentafluoropropionic acid. Elution was performed with a gradient of 33–66% at 1 mL/min flow rate during 30 min. Relevant fractions were collected and analyzed on a 20% homogenous Phast Gel (Amersham Pharmacia Biotech, Uppsala, Sweden) and pure samples were lyophilized.
Purification of the triple mutants ABD (E3A, V6A, L7A), ABD (R10A, E11A, K14A), and ABD (K35A, I38A, D39A) was done by HSA affinity chromatography, as described by Nygren et al. (1988).
Protein concentration measurements
Protein concentration was determined by three independent methods: Bradford quantification (Bradford 1976); measuring the absorbance at 280 nm using the specific absorption constant of 0.315–0.375 [g/L] (dependent on the mutant) as described by Gill and von Hippel (1989); and estimation from SDS-PAGE. To further verify the protein concentration, two samples of Z-ABD were analyzed by amino acid analysis.
CD spectroscopy analysis
Protein samples were dissolved in a potassium phosphate buffer (8.1 mM K2HPO4, 1.9 mM KH2PO4 at pH 7.5) to a concentration of ∼10 μM and filtered. Data were collected using a Jasco J-720 spectropolarimeter (Jasco, Japan). The scanning speed was 10 nm/min and data were collected from 260 to 190 nm. The cell path length was 1 mm and the protein concentration, 10 μM. Accurate protein concentration was determined by amino acid analysis and values of three measurements were averaged for each protein analyzed. CD spectra for the mutants were obtained by subtracting the signals for the Z domain, after adjustment for differences in protein concentration. The subtraction was followed by normalization for amino acid content (Nord et al. 1997).
Biospecific interaction analysis
Affinity blotting
Samples were separated on an SDS PAGE in the Phast System (Amersham Pharmacia Biotech, Uppsala, Sweden) and transferred by thermal blotting over 30 min at 70°C to a nitrocellulose membrane (BioRad Laboratories). After blocking with 1% casein (Semper AB, Sweden) in PBS for 30 min (0.1 M NaCl, 33 mM Na2HPO4, 17 mM NaH2PO4•2H2O at pH 7.2) the membrane was incubated with 50 μg of biotinylated HSA (Pharmacia Corp., Stockholm, Sweden) in 15 mL of PBST (PBS supplemented with 0.05% Tween 20) for another 30 min. (The biotinylation was performed according to manufacturer's recommendations.) After washing four times with PBST, the membrane was incubated with a streptavidin–alkaline phosphatase conjugate, 7 μL in 15 mL of PBST (Boeringer Mannheim, Germany). Finally, the membrane was incubated with BCIP/NBT (5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium; Sigma), a substrate for alkaline phosphatase, according to the manufacturer's recommendation.
Biacore analysis
Lyophilized protein samples were dissolved in 1*HBS (10 mM HEPES at pH 7.4, 150 mM NaCl, 3.4 mM EDTA, 0.5 % surfactant P 20) and filtered. A Biacore 2000 instrument (Biacore AB, Sweden) was used for real-time biospecific interaction analysis. HSA and human IgG (Pharmacia Corp., Stockholm, Sweden) were immobilized on the surface of the CM 5 chip (Biacore AB), according to Biacore's recommendations. The immobilization resulted in ∼3000 resonance units (RU). To compare the affinity between the different variants, a concentration of 100 nM of each protein was analyzed. To assure the differences in affinity between the mutants, the average values were calculated from at least two measurements. The signal from a nonimmobilized surface was subtracted.
Furthermore, a kinetic study was carried out. Ten different concentrations were used for each variant to have comparable responses on the Biacore surface. Ten samples for the ABD variants E3A, D19A, N23A, L24A, E32A, and the parental ABD were prepared in concentrations ranging from 20 to 200 nM. For S18A and K22A, the concentrations were in the range of 100 to 300 nM. For Y20A, Y20A, E32A, and also for Y20A, E40A, sample concentration was from 220 to 400 nM. When analyzing the affinity of Y21A, the concentrations were from 50 to 450 nM. For mutant S18A, Y20A, K22A, the concentrations were 100 to 1000 nM, and finally for K29A, 60 to 240 nM. At least four independent samples were analyzed for all concentrations. The evaluations of the results were done by the use of BIAevaluation 3.02 software (Biacore AB). From calculated kon and koff values, Kaff, KD, and also the change in free binding energy (δδG) was deduced for each ZABD variant.
Acknowledgments
We thank Mats Wikström (Pharmacia Corp.) for allowing us to use the CD-spectropolarimeter and Peter Nilsson for valuable discussions concerning the BIACORE data.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.02802.
References
- Albeck, S. and Schreiber, G. 1999. Biophysical characterization of the interaction of the beta-lactamase TEM-1 with its protein inhibitor BLIP. Biochemistry 38 11–21. [DOI] [PubMed] [Google Scholar]
- Albeck, S., Unger, R., and Schreiber, G. 2000. Evaluation of direct and cooperative contributions towards the strength of buried hydrogen bonds and salt bridges. J. Mol. Biol. 298 503–520. [DOI] [PubMed] [Google Scholar]
- Bass, S.H., Mulkerrin, M.G., and Wells, J.A. 1991. A systematic mutational analysis of hormone-binding determinants in the human growth hormone receptor. Proc. Natl. Acad. Sci. 88 4498–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Carter, D.C., and He, X.M. 1990. Structure of human serum albumin. Science 249 302–303. [DOI] [PubMed] [Google Scholar]
- Clackson, T., Ultsch, M.H., Wells, J.A., and de Vos, A.M. 1998. Structural and functional analysis of the 1:1 growth hormone:receptor complex reveals the molecular basis for receptor affinity. J. Mol. Biol. 277 1111–1128. [DOI] [PubMed] [Google Scholar]
- Cunningham, B.C. and Wells, J.A. 1993. Comparison of a structural and a functional epitope [published erratum appears in J. Mol. Biol. 1994;8;237:513]. J. Mol. Biol. 234 554–563. [DOI] [PubMed] [Google Scholar]
- Curry, S., Mandelkow, H., Brick, P., and Franks, N. 1998. Crystal structure of human serum albumin complexed with fatty acid reveals an asymmetric distribution of binding sites [see comments]. Nat. Struct. Biol. 5 827–835. [DOI] [PubMed] [Google Scholar]
- Forsgren, A. and Sjöquist, J. 1966. "Protein A" from S. aureus. I. Pseudo-immune reaction with human gamma-globulin. J. Immunol. 97 822–827. [PubMed] [Google Scholar]
- Gårdsvoll, H., Dano, K., and Ploug, M. 1999. Mapping part of the functional epitope for ligand binding on the receptor for urokinase-type plasminogen activator by site-directed mutagenesis. J. Biol. Chem. 274 37995–38003. [DOI] [PubMed] [Google Scholar]
- Gill, S.C. and von Hippel, P.H. 1989. Calculation of protein extinction coefficients from amino acid sequence data [published erratum appears in Anal. Biochem. 1990;189:283]. Anal. Biochem. 182 319–326. [DOI] [PubMed] [Google Scholar]
- Gülich, S., Linhult, M., Nygren, P.Å., Uhlén, M., and Hober, S. 2000. Stability towards alkaline conditions can be engineered into a protein ligand [In Process Citation]. J. Biotechnol. 80 169–178. [DOI] [PubMed] [Google Scholar]
- Hammarberg, B., Nygren, P.Å., Holmgren, E., Elmblad, A., Tally, M., Hellman, U., Moks, T., and Uhlén, M. 1989. Dual affinity fusion approach and its use to express recombinant human insulin-like growth factor II. Proc. Natl. Acad. Sci. 86 4367–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jendeberg, L., Persson, B., Andersson, R., Karlsson, R., Uhlén, M., and Nilsson, B. 1995. Kinetic analysis of the interaction between protein A domain variants and human Fc using plasmon resonance detection. J. Mol. Recognit. 8 270–278. [DOI] [PubMed] [Google Scholar]
- Johansson, M.U., de Chateau, M., Wikström, M., Forsén, S., Drakenberg, T., and Björck, L. 1997. Solution structure of the albumin-binding GA module: a versatile bacterial protein domain. J. Mol. Biol. 266 859–865. [DOI] [PubMed] [Google Scholar]
- Johnson, W.C. Jr. 1990. Protein secondary structure and circular dichroism: A practical guide. Proteins 7 205–214. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J., Jonasson, P., Nygren, P.Å Uhlén, M., Jendeberg, L., Nilsson, B., and Kördel, J. 1996. The serum albumin-binding domain of streptococcal protein G is a three-helical bundle: a heteronuclear NMR study. FEBS Lett. 378 190–194. [DOI] [PubMed] [Google Scholar]
- Kronvall, G., Simmons, A., Myhre, E.B., and Jonsson, S. 1979. Specific absorption of human serum albumin, immunoglobulin A, and immunoglobulin G with selected strains of group A and G streptococci. Infect. Immunol. 25 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libon, C., Corvaia, N., Haeuw, J.F., Nguyen, T.N., Ståhl, S., Bonnefoy, J.Y., and Andreoni, C. 1999. The serum albumin-binding region of streptococcal protein G (BB) potentiates the immunogenicity of the G130–230 RSV-A protein. Vaccine 17 406–414. [DOI] [PubMed] [Google Scholar]
- Lottenberg, R., Broder, C.C., and Boyle, M.D. 1987. Identification of a specific receptor for plasmin on a group A streptococcus. Infect. Immunol. 55 1914–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makrides, S.C., Nygren, P.Å., Andrews, B., Ford, P.J., Evans, K.S., Hayman, E.G., Adari, H., Uhlén, M., and Toth, C.A. 1996. Extended in vivo half-life of human soluble complement receptor type 1 fused to a serum albumin-binding receptor. J. Pharmacol. Exp. Ther. 277 534–542. [PubMed] [Google Scholar]
- Nilsson, B., Moks, T., Jansson, B., Abrahmsen, L., Elmblad, A., Holmgren, E., Henrichson, C., Jones, T.A., and Uhlén, M. 1987. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. 1 107–113. [DOI] [PubMed] [Google Scholar]
- Nord, K., Gunneriusson, E., Ringdahl, J., Ståhl, S., Uhlén, M., and Nygren, P.Å. 1997. Binding proteins selected from combinatorial libraries of an alpha-helical bacterial receptor domain. Nature Biotechnol. 15 772–777. [DOI] [PubMed] [Google Scholar]
- Nygren, P.Å., Eliasson, M., Abrahmsen, L., Uhlén, M., and Palmcrantz, E. 1988. Analysis and use of the serum albumin binding domains of streptococcal protein G. J. Mol. Recognit. 1 69–74. [DOI] [PubMed] [Google Scholar]
- Nygren P.Å, Per Flodby, M.U., Andersson, R., and Wigzell, H. 1991. In vivo stabilization of a human recombinant CD4– derivate by fusion to a serum-albumin-binding receptor. Vaccines 96 363–368. [Google Scholar]
- Olsson, A., Eliasson, M., Guss, B., Nilsson, B., Hellman, U., Lindberg, M., and Uhlén, M. 1987. Structure and evolution of the repetitive gene encoding streptococcal protein G. Eur. J. Biochem. 168 319–324. [DOI] [PubMed] [Google Scholar]
- Otzen, D.E. and Fersht, A.R. 1999. Analysis of protein–protein interactions by mutagenesis: Direct versus indirect effects. Protein Eng. 12 41–45. [DOI] [PubMed] [Google Scholar]
- Schreiber, G. and Fersht, A.R. 1995. Energetics of protein–protein interactions: Analysis of the barnase–barstar interface by single mutations and double mutant cycles. J. Mol. Biol. 248 478–486. [DOI] [PubMed] [Google Scholar]
- Sjölander, A., Nygren, P.Å. Ståhl, S., Berzins, K. Uhlén, M. Perlmann, P., and Andersson, R. 1997. The serum albumin-binding region of streptococcal protein G: A bacterial fusion partner with carrier-related properties. J. Immunol. Methods 201 115–123. [DOI] [PubMed] [Google Scholar]
- Ståhl, S. and Nygren, P.Å. 1999. The use of gene fusions to protein A and protein G in immunology and biotechnology. Path. Bio. 45 66–76. [PubMed] [Google Scholar]
- Vaughan, C.K., Buckle, A.M., and Fersht, A.R. 1999. Structural response to mutation at a protein–protein interface. J. Mol. Biol. 286 1487–1506. [DOI] [PubMed] [Google Scholar]