Fig. 2.
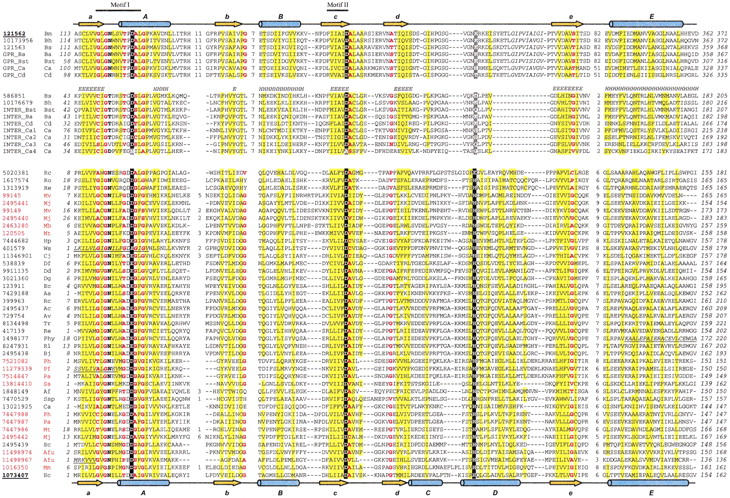
Multiple sequence alignment of the GPR/hydrogenase maturation protease superfamily. Groups of sequences are separated by empty lines. GPRs and hydrogenase maturation proteases are the top and the bottom groups, respectively, with the intermediate sequences in between. Each sequence is identified by the NCBI gene identification number (gi), except for those obtained from the ERGO database or NCBI unfinished genomes DNA databases, which use the group name abbreviations followed by the species abbreviations. Sequence identifiers are followed by abbreviations of the species names. The gi numbers of the sequences with known structures are underlined and in bold. The archaeal identifiers are marked with red. The first and the last residue numbers of the sequences are indicated, and the total length of the proteins is shown at the end (italicized numbers). Residues in unconserved loop regions are not displayed, with the number of omitted residues shown instead. The metal-binding residues are shown in white bold letters on black background. The putative third metal ligand in the GPRs and intermediate sequences is shaded in gray. Uncharged residues at mainly hydrophobic positions are shaded yellow. Conserved small residues (G, A, S, C, T, P, V, D, and N) are in red bold letters. Sequences with apparently incorrect starting or ending positions are corrected, with the added N-terminal fragments in underlined italicized letters. One loop that is not modeled in the structure of GPR is shown in italicized letters. Diagrams of the secondary structural elements are shown at the top (GPR: 1c8b, chain A) and bottom (HybD: 1cfz, chain A). β-strands and α-helices are shown as yellow arrows and blue cylinders, respectively. The labeling associated with the secondary structure elements corresponds to Figure 1 ▶. Predicted secondary structure by Jpred2 is shown for the intermediate sequences (E indicates a strand; H indicates a helix). The species name abbreviations are: Aa, Aquifex aeolicus; Ac, Azotobacter chroococcum; Af, Acetomicrobium flavidum; Afu, Archaeoglobus fulgidus; Av, Azotobacter vinelandii; Ba, Bacillus anthracis; Bh, Bacillus halodurans; Bj, Bradyrhizobium japonicum; Bm, Bacillus megaterium; Bs, Bacillus subtilis; Bst, Bacillus stearothermophilus; Ca, Clostridium acetobutilycum; Cd, Clostridium difficile; Cj, Campylobacter jejuni; Dd Desutobacterium dehalogenans; Df, Desulfovibrio fructosovorans; Dg, Desulfovibrio gigas; Ec, Escherichia coli; Hp, Helicobacter pylori; Mb, Methanosarcina barkeri; Mj, Methanococcus jannaschii; Mm, Methanosarcina mazei; Mt, Methobacterium thermoautotrophicum; Mv, Methanococcus voltae; Pa, Pyrococcus abyssi; Pf, Pyrococcus furiosus; Ph, Pyrococcus horikoshii; Phy, Pseudomonas hydrogenovora; Rc, Rhodobacter capsulatus; Re, Ralstonia eutropha; Rl, Rhizobium leguminosarum; Ro, Rhodococcus opacus; Ss, Sulfolobus solfataricus; Ssp, Synechocystis sp.; Tr, Thiocapsa roseopersicina; Ws, Wolinella succinogenes. The alignment was generated using T-Coffee (Notredame et al. 2000), followed by manual adjustment.
