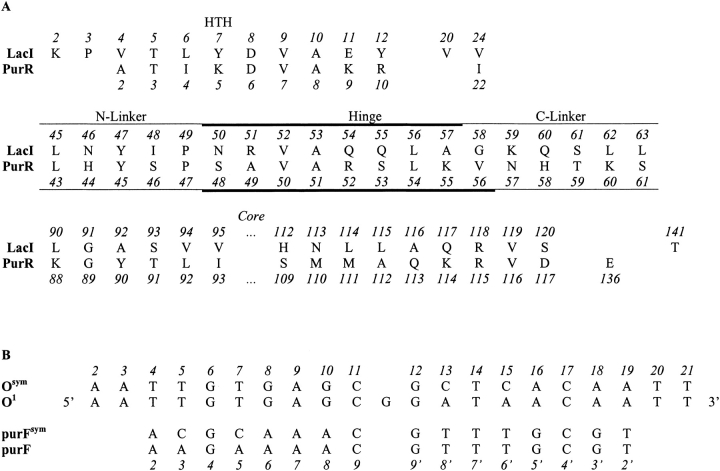
Fig. 2.
(A) LacI and PurR sequence alignments. Residue numbers for LacI are above the one-letter amino acid code; numbers for PurR are below. Structural regions are denoted above the LacI residue numbers. Sites in the N- and C-linkers are denoted with a thin line, and those in the hinge helix are indicated with a bold line. Note that the hinge helix of PurR extends one residue further than that of LacI. The sequences of LacI 141 and PurR 136, which do not align with each other, are also included because these residues make contacts to the N-linker regions. Intervening residues are omitted for clarity. (B) Sequences of LacI and PurR operators. DNA bases are denoted with the one-letter code. LacI binds both the natural operator O1 and an "optimized" sequence Osym with high affinity (Gilbert and Maxam 1973; Sadler et al. 1983; Simons et al. 1984). PurR binds both purF and a palindromic form of that operator (purFsym; Schumacher et al. 1994; Makaroff and Zalkin 1985). LacI crystal structures are in complex with Osym (Bell and Lewis 2000), whereas the PurR structure 1wet was determined in complex with purF (Schumacher et al. 1997). However, this structure must contain some disorder, since the unit cell only contains one purR monomer and one DNA strand even though purF is asymmetric (Schumacher et al. 1997). The NMR structure of LacI(1–62) is also in complex with Osym (1cjg; Spronk et al. 1999a). Numbers at the top of the figure are those of Lac Osym from the pdb file 1efa and are used throughout this report. Numbers at the bottom of this alignment correspond to the base pair numbers used by Schumacher et al. (1994) for PurR structural descriptions.