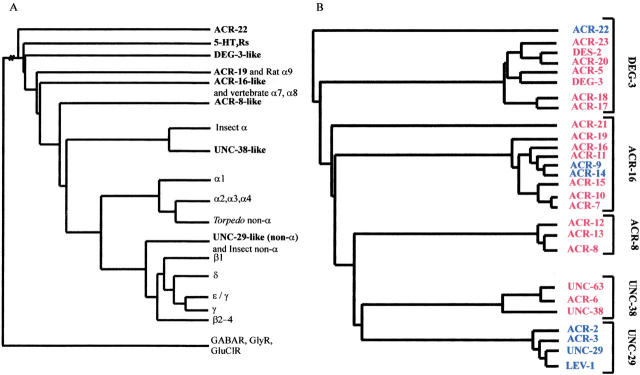
Fig. 2.
(A) An alignment of all nAChR subunit sequences available in public databases and a selection of anionic selective GABAR, GlyR, and GluClR was generated using default parameters of ClustalW (Thompson et al. 1994). The anionic channels were selected as the out-group to root the tree. The global tree was constructed using the Protdist (Dayhoff PAM algorithm) and Neighbor programs of the PHYLIP package. For reasons of clarity, related groups of vertebrate, insect, and C. elegans subunits groups are illustrated by single branches in A. The insect branch includes the two available Myzus persicae sequences (X81887, X81888) in addition to the Manduca sexta (Y09795), Locusta migratoria (AJ000390, AJ000391, AJ000393), Heliothis virescens (AJ000399) and Drosophila melanogaster ALS (X07194), SBD (X55676) and Dα3 (Y15594) subunits. UNC-38-like homologs have been isolated from the parasitic nematodes Ascaris suum (AJ011382), Trichostrongylus columbriformis (U56903), Haemonchus contortus (U72490), and a non-α subunit, which is more similar to UNC-38 than other non-α subunits, from Onchocerca volvulus (L20465). (B) This tree shows the C. elegans groups expanded to reveal all C. elegans nAChR subunits. Five distinct groups of C. elegans subunits can be recognized: four groups of mainly α subunits; DEG-3-like, ACR-16-like, UNC-38-like, and ACR-8-like nAChR α subunits, in addition to the UNC-29-like group containing only non-α subunits. The alignment and global tree used to generate this Fig. are available on request from the authors.