Fig. 2.
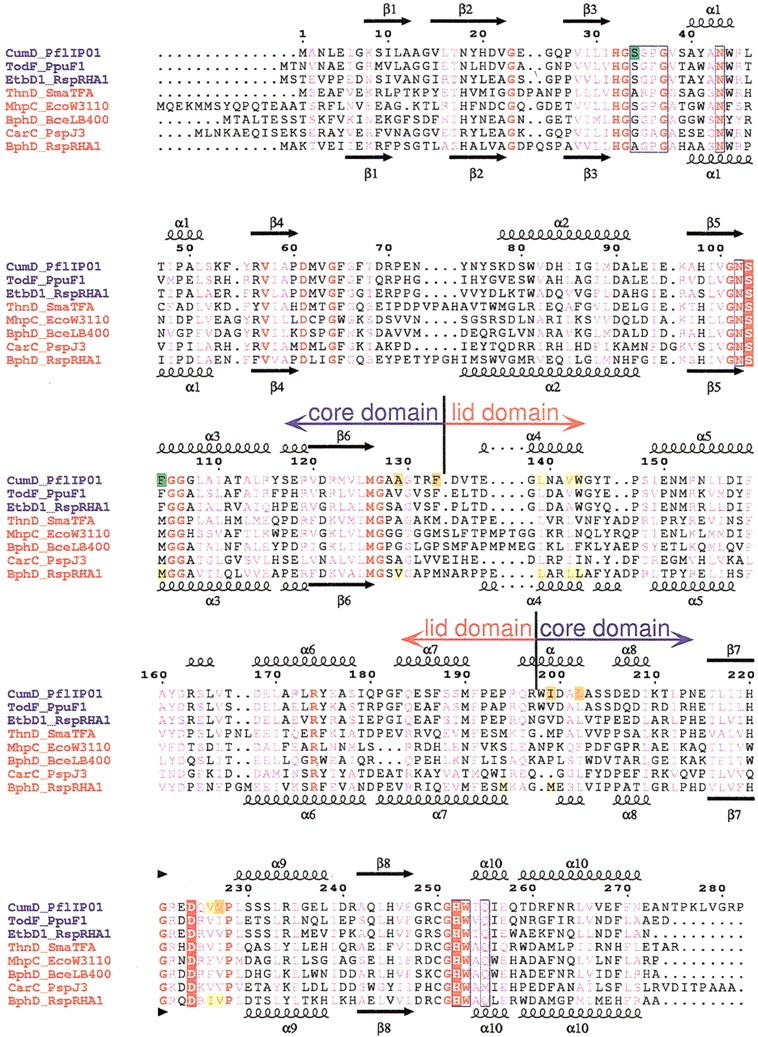
Sequence alignment of HODA hydrolases. Multiple sequence alignment was performed using program ClustalX (Thompson et al. 1997) and then modified on the basis of structural alignment. The sequence names are given in blue and red for members of the monoalkylbenzene and biphenyl groups, respectively. The numbering above the alignment is for the CumD sequence. The secondary structures and their designations are shown above CumD and below RHA1 BphD; arrows, large coils, and small coils represent the β-strands, α-helices, and 310 helices, respectively. Completely conserved residues among the presented sequences are colored red, and conserved residues are shown in pink. The catalytic residues are indicated by red rectangles. The residues involved in the recognition of the 2-hydroxy-6-oxohexa-2,4-dienoate group of the substrate are surrounded by blue frames. The residues involved in the recognition of the carboxylate group and isopropyl group and in the formation of the deeper space of the D-part of CumD are indicated by green, yellow, and orange rectangles, respectively. The residues involved in the recognition of the formation of the D-part of RHA1 BphD are indicated by yellow rectangles. CumD from Pseudomonas fluorescens IP01 (D83955), TodF from P. putida F1 (Y18245), EtbD1 from Rhodococcus sp. RHA1 (AB004320), ThnD from Sphingomonas macrogoltabidus TFA (AF204963), MhpC from Escherichia coli W3110 (D86239), BphD from Burkholderia cepacia LB400 (X66123), CarC from Acrobacterium sp. strain J3 (K. Morii, H. Habe, H. Nojiri, and T. Omori, unpubl.), and BphD from Rhodococcus sp. strain RHA1 (D88016).
