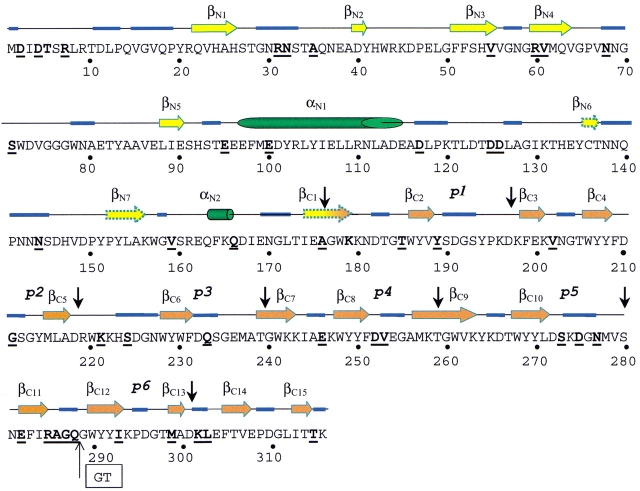
Fig. 4.
Predicted secondary structure of Ejl. The arrows (β-strand), cylinders (α-helix), and blue traces (loops) correspond to those amino acid residues whose conformation is predicted with accuracy higher than 85.7% by at least two of the methods employed (PHD, PSIpred, and the Jpred servers); discontinuous arrows indicate three β-strands predicted with lower reliability. Vertical black arrows indicate the sequence region comprising the sequence repeats (p1–p6) of the ChBM. Bold letters indicate the Ejl amino acid muted in relation to LytA amidase. The "GT" inset indicates the position of the double deletion in the p6 repeat of Ejl.