Fig. 1.
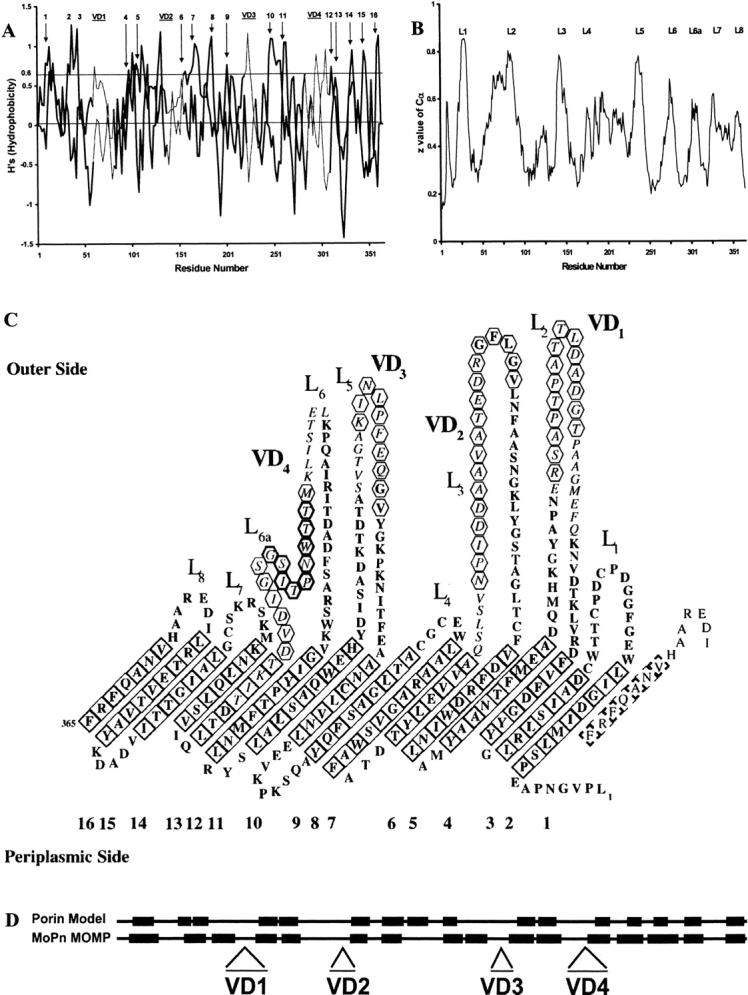
(A) β-side hydrophobicity plot of Chlamydia trachomatis mouse pneumonitis (MoPn) MOMP. Values for even- and odd-numbered residues are plotted as two separate curves. Peaks that have been assigned to transmembrane β-strands in panel C are labeled 1 to 16. The locations of the four variable domains (VD1 through VD4) are indicated by bars at the top of the plot, and the residues corresponding to those regions are joined by thin lines. (B) Topology plot from an artificial neural network program trained on bacterial outer-membrane proteins: predicted z-values are plotted against the C. trachomatis MoPn MOMP amino-acid sequence. The extracellular loops are marked L1 through L8 as assigned in panel C. (C) Predicted topology of the MOMP of C. trachomatis MoPn. The 16-stranded β-barrel has been unrolled and the view is from its outer surface. Italics highlight amino acids in the VDs, while tilted squares indicate residues in transmembrane β-strands, with a bold outline if the side chains are facing away from the pore. Antibodies have been mapped to the segments indicated by hexagons. The conserved peptide in VD4 is indicated by bold hexagons. (The amino-acid sequence is in one-letter code.) (D) Secondary-structure homology using FORESST. The predicted membrane-spanning β-sheets (black boxes) of C. trachomatis MoPn MOMP more closely align with those of the FORESST general porin model after MOMP segments containing the four VDs were deleted.
