Abstract
Phenylalanine hydroxylase (PAH) is activated by its substrate phenylalanine, and through phosphorylation by cAMP-dependent protein kinase at Ser16 in the N-terminal autoregulatory sequence of the enzyme. The crystal structures of phosphorylated and unphosphorylated forms of the enzyme showed that, in the absence of phenylalanine, in both cases the N-terminal 18 residues including the phosphorylation site contained no interpretable electron density. We used nuclear magnetic resonance (NMR) spectroscopy to characterize this N-terminal region of the molecule in different stages of the regulatory pathway. A number of sharp resonances are observed in PAH with an intact N-terminal region, but no sharp resonances are present in a truncation mutant lacking the N-terminal 29 residues. The N-terminal sequence therefore represents a mobile flexible region of the molecule. The resonances become weaker after the addition of phenylalanine, indicating a loss of mobility. The peptides corresponding to residues 2–20 of PAH have different structural characteristics in the phosphorylated and unphosphorylated forms, with the former showing increased secondary structure. Our results support the model whereby upon phenylalanine binding, the mobile N-terminal 18 residues of PAH associate with the folded core of the molecule; phosphorylation may facilitate this interaction.
Keywords: Autoregulatory sequence, mutagenesis, nuclear magnetic resonance (NMR), phenylalanine hydroxylase, phosphorylation
Phenylalanine hydroxylase (PAH) is a metabolic enzyme that converts phenylalanine to tyrosine using molecular oxygen, enzyme-bound iron, and a 6R-tetrahydrobiopterin (BH4) cofactor (Kaufman 1993; Hufton et al. 1995; Kappock and Caradonna 1996; Fitzpatrick 2000). PAH is a member of the aromatic amino acid hydroxylase family, together with tyrosine hydroxylase (TH) and tryptophan hydroxylase (TPH). TH and TPH are involved in the biosynthesis of the neurotransmitters, dihydroxyphenylalanine (l-DOPA) and serotonin, respectively. The aromatic amino acid hydroxylases share a similar enzyme mechanism, and have a common three-domain structure consisting of an N-terminal regulatory domain, a catalytic domain, and a C-terminal tetramerization domain; the highest sequence and structural similarity is found in the catalytic domain (Flatmark and Stevens 1999). Over 300 different mutations in the PAH gene have been found to be associated with the disease phenylketonuria (PKU) (Nowacki et al. 1997).
The regulatory domains of the aromatic amino acid hydroxylases are not highly conserved, reflecting the different modes of regulation required by these enzymes. PAH controls the level of phenylalanine, an essential amino acid, which is subject to large fluctuations due to dietary intake. On the one hand, the levels of PAH in the liver are such that if uncontrolled, the enzyme would rapidly deplete the phenylalanine stores; on the other hand, the metabolites of phenylalanine are toxic to the developing brain. PAH is therefore tightly controlled by a variety of mechanisms. The major regulatory mechanisms include activation by phenylalanine, inhibition by BH4, and additional activation by phosphorylation (Hufton et al. 1995). Activation by the substrate phenylalanine is considered the major regulatory event; phenylalanine binds to a proposed allosteric site on PAH (here referred to as the activation site), and induces large conformational changes (Shiman et al. 1979; Hufton et al. 1995; Fitzpatrick 2000). The natural cofactor tetrahydrobiopterin (BH4) acts as a negative regulator, by blocking phenylalanine activation (Shiman and Gray 1980; Hufton et al. 1995; Fitzpatrick 2000).
cAMP-dependent protein kinase phosphorylates PAH at Ser16 (Wretborn et al. 1980). Although phenylalanine is the overriding factor in activation of PAH, phosphorylation acts as a mediator of phenylalanine activation by decreasing the phenylalanine concentration required to activate the enzyme (Shiman et al. 1982; Doskeland et al. 1984; Hufton et al. 1995; Fitzpatrick 2000). Thirty to 40% less phenylalanine is required for the half-maximal activation of the phosphorylated enzyme when compared to the unphosphorylated protein (Doskeland et al. 1984). Substitution of Ser16 with negatively charged amino acids, glutamate, or aspartate results in activation of the enzyme (Citron et al. 1994; Kowlessur et al. 1995). The phosphorylation at Ser16 is considered a physiologically relevant regulatory mechanism of PAH (Donlon and Kaufman 1978).
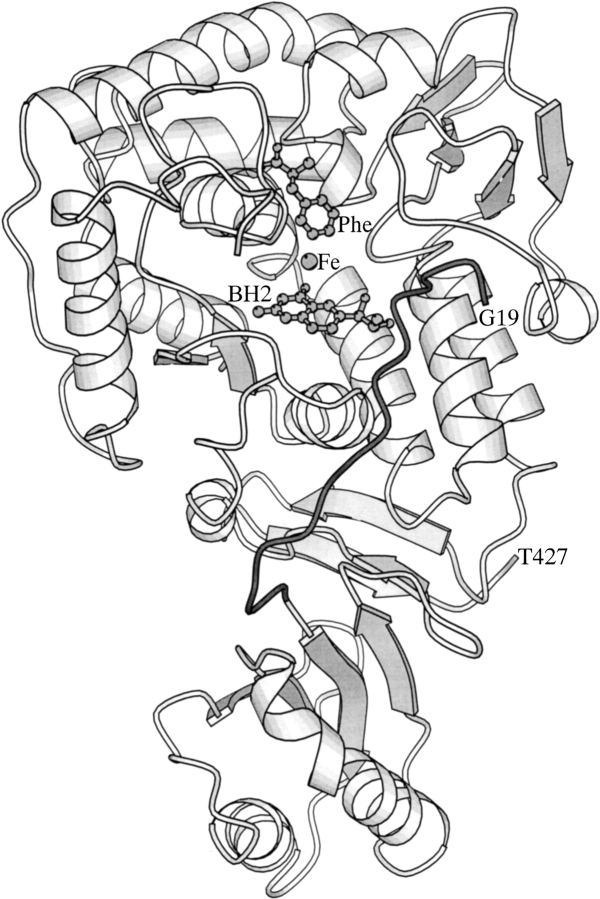
Several crystal structures of fragments of PAH and TH have recently been determined (Flatmark and Stevens 1999). The crystal structure of the dimeric rat PAH lacking the C-terminal, 24-residue tetramerization domain (PAH1–428) represents the only available structure of an aromatic amino acid hydroxylase that contains the regulatory domain (Kobe et al. 1999). This structure revealed that the N-terminal sequence comprising amino acids 19–29 reaches into the active site of the catalytic domain (Fig. 1 ▶), acting as an intrasteric autoregulatory sequence (Kobe et al. 1999; Kobe and Kemp 1999; Jennings et al. 2001; Wang et al. 2001). Remarkably, the structures of both the phosphorylated and unphosphorylated forms of PAH1–428 were identical, with no electron density evident for the 18 N-terminal residues containing the phosphorylation site Ser16 (Kobe et al. 1999).
Fig. 1.
A ribbon diagram of the crystal structure of phosphorylated PAH1–428 monomer (Protein Data Bank code 1PHZ; Kobe et al. 1999) is shown with the autoregulatory region (residues Gly19 to Gln30) in dark gray. For orientation purposes only, the proposed positions of the substrate phenylalanine (Phe), and the pterin inhibitor, 7,8-dihydrobiopterin (BH2) are shown based on the model of Teigen et al. (1999).
To elucidate the structural basis of PAH activation, we here complement the crystallographic results with the method of nuclear magnetic resonance (NMR). It has previously been demonstrated that NMR can be used to characterize mobile regions of larger proteins that would not ordinarily be expected to yield useful information in an NMR experiment (Landry et al. 1993). We show here that the NMR spectrum of the dimeric PAH1–428 (∼98 kD) contains sharp peaks, corresponding to the N-terminal region of the molecule, superimposed on an envelope of broad, overlapping resonances. The addition of phenylalanine resulted in a decrease of the intensity of the peaks. The NMR spectrum of the peptide corresponding to residues 2–20 of PAH revealed a stabilization of secondary structure in the phosphorylated form when compared to the unphosphorylated form. Our results support the model for regulation of PAH activity whereby the mobile N-terminal 18 residues of PAH bind to the folded core of the molecule upon the addition of phenylalanine, with phosphorylation facilitating this interaction.
Results
Assignment of resonances in the N-terminal sequence of PAH1–428
PAH1–428 is a dimeric protein of ∼98 kD. In this form and under normal solution conditions for NMR spectroscopy, crosspeaks in two-dimensional 1H homonuclear spectra would not be expected to be observed. The NH to aliphatic region of the two-dimensional 1H,1H TOCSY spectra of phosphorylated PAH1–428 (Fig. 2A ▶), however, contains crosspeaks of approximately 25 spin systems indicating that there are regions within the molecule that are substantially more mobile. The crystal structure of PAH1–428 (Kobe et al. 1999) suggests that the N-terminal region is disordered; we therefore acquired the analogous spectra of PAH lacking the first 29 residues (PAH30–428). The TOCSY spectra of PAH30–428 (data not shown) do not show any crosspeaks between NH to aliphatic residues, supporting the conclusion that the N-terminal region of PAH1–428 is significantly mobile.
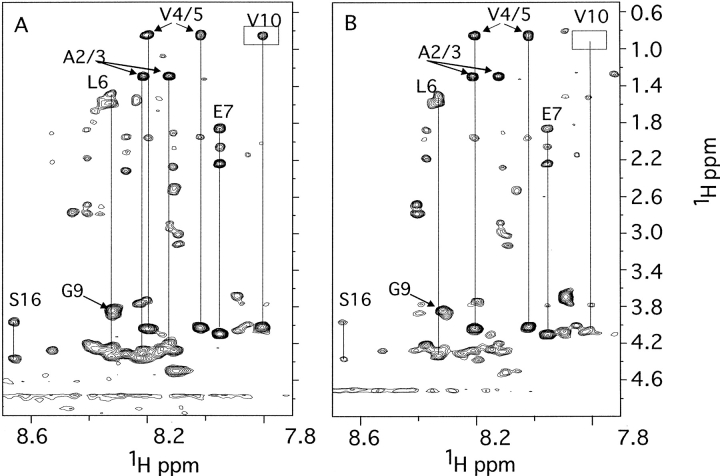
Fig. 2.
The NH to aliphatic region of the 2D 1H-1H TOCSY spectrum of ∼0.1 mM phosphorylated PAH1–428 without phenylalanine (A) and with 200 mM phenylalanine (B). Assigned spin systems are indicated.
In the TOCSY spectrum of phosphorylated PAH1–428 (Fig. 2A ▶), a number of spin system topologies can be identified corresponding to residues in the N-terminal region. Three Val, two Ala, two Ser, one Gly, one Glu or Gln, three AMX systems (characteristic of residues such as Asn and Phe), and several "long-chain" systems typical of Leu, Lys, or Arg are identifiable. 2D NOESY spectra of PAH1–428 were also acquired, and while corresponding intraresidue NOEs are observed, no unambiguous sequential NOE assignments could be identified, partly due to overlapping and broad crosspeaks from the bulk of the protein.
To obtain further data to assist the assignment of these residues we acquired TOCSY and NOESY spectra of a synthetic peptide corresponding to residues 2–20 of PAH1–428 (Fig. 3 ▶). TOCSY spectra of the phosphorylated peptide PAH2–20 showed 23 spin systems, indicating there was some degree of heterogeneity. However, the chemical shifts of several systems are extremely similar to those present in PAH1–428. The most up-field NH at 7.78 ppm shows characteristic connectivities of a Val residue, and the NOESY spectrum shows NH to NH and NH to CαH NOEs to a Gly residue and to a Leu residue, assigning these residues to Gly9, Val10, Leu11 (there is only one Gly-Val-Leu sequence motif present in PAH; peptide numbering is according to the PAH sequence; therefore, the first peptide residue is numbered 2). The NH to NH NOEs for this region show that the peptide spends some of the time in a turn-like structure, and as the chemical shifts are similar in the spectra of PAH1–428, this conformation may be conserved. The most down-field NOEs at 8.53 and 8.66 ppm show two NH-CHα–CHβ systems with chemical shifts typical of Ser. Comparison to the nonphosphorylated peptide shows that both of these systems are absent, indicating they belong to Ser16, and that the residue is in two distinct conformations. As the most downfield system (8.66 ppm) in PAH1–428 also appears to be a Ser, we have assigned this system to the phosphorylated Ser16. Further support for this assignment was provided by the absence of an analogous downfield signal in spectra of unphosphorylated PAH1–428 (data not shown). In these spectra resonances at similar chemical shifts to phosphorylated PAH1–428 were observed for three Val, two Ala, Gly, and a possible Glu.
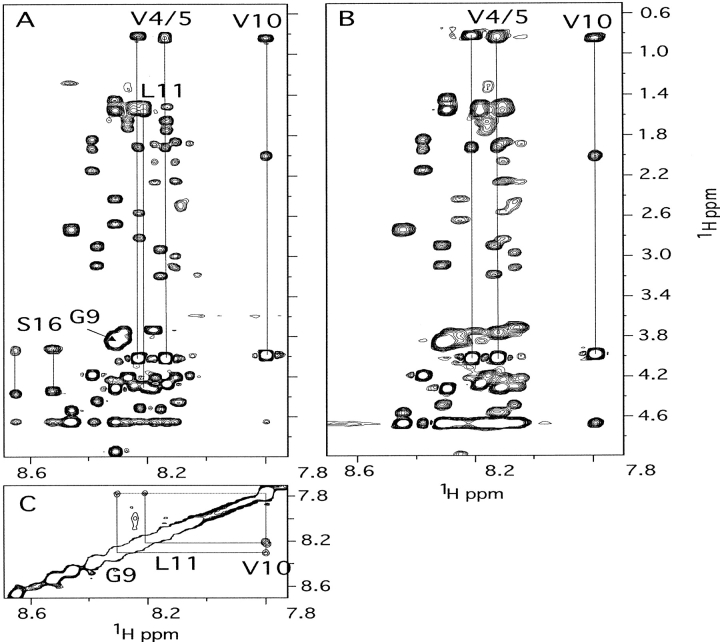
Fig. 3.
The NH to aliphatic region of the 2D 1H-1H TOCSY spectrum of phosphorylated (A) and unphosphorylated (B) peptide PAH2–20 and the NH to NH region of the 1H-1H NOESY spectrum of phosphorylated (C) peptide PAH2–20. Assigned spin systems are indicated.
Effect of phenylalanine activation of PAH as assessed by NMR
The addition of phenylalanine (200 mM) to the samples of phosphorylated PAH1–428 resulted in a decrease in intensity of a number of sharp resonances (Fig. 2B ▶). Most notably, the resolved resonances of Val10 and Ser16 are clearly attenuated, but to different extents. The signals near 8.1 and 8.3 ppm also decrease in intensity with the addition of phenylalanine. On the other hand, resonances that are assigned to Ala2, Ala3, Val4, Val5, and tentatively to Glu7 do not appear attenuated. Similar attenuation of signals was observed for unphosphorylated PAH1–428 (data not shown; for technical reasons, we were unable to follow the effect of increasing concentrations of phenylalanine on phosphorylated and unphosphorylated enzyme). Addition of phenylalanine to PAH2–20 had no effect on the spectra; we therefore conclude that the effects observed on PAH1–428 are due to phenylalanine binding and not due to dynamic range or other spurious effects.
Discussion
PAH is activated by its substrate phenylalanine, and through phosphorylation by cAMP-dependent protein kinase at Ser16 in the N-terminal autoregulatory sequence of the enzyme. The crystal structure of PAH1–428 (Kobe et al. 1999) and deletion mutagenesis (Jennings et al. 2001; Wang et al. 2001) revealed an autoinhibitory role for the N-terminal 29 residues of the enzyme. Intriguingly, no interpretable electron density was observed for the residues 1–18 containing the phosphorylation site (residue Ser16), in both the phosphorylated and unphosphorylated forms. The structural basis of the activation by phenylalanine and phosphorylation therefore remained elusive.
In the present study, we used NMR spectroscopy to address some questions not amenable to crystallographic analysis, in particular the structural behavior of the N-terminal sequence of PAH in the different stages of the regulatory pathway. In a context of a large protein such as PAH, only the mobile regions were expected to produce sharp peaks in the NMR spectra, overlayed onto the background of broad overlapping resonances. A similar approach has previously been used to study the mobile regions of the cochaperonin GroES (Landry et al. 1993, 1996). Although sequential assignment proved difficult in our system, we confirmed that the N-terminal sequence was responsible for the sharp resonances, based on the following evidence: (1) the peaks were present in the spectra of PAH1–428, but not PAH30–428; (2) several spin systems present in the N-terminal 18 residues of the protein could be identified in the spectrum; (3) a serine spin system considerably shifted downfield was present in the phosphorylated sample; and (4) a serine spin system with similar characteristics was present in the phosphorylated peptide PAH2–20, but not in the unphosphorylated peptide.
Studies with the peptide PAH2–20 indicated an influence of phosphorylation upon solution behavior; the addition of the phosphate group resulted in a higher degree of secondary structure. Although the consequences, within the context of the enzyme, of the effect of the phosphate group on the structure and dynamics of the local sequence are not clear, this effect may play an important role in the mechanism of activation of PAH activity by phosphorylation.
In PAH1–428, it is more difficult to determine the specific effects of phosphorylation on the N-terminal sequence for two reasons: (1) the concentration of the protein is lower to prevent aggregation, limiting the observable signals; and (2), the tethering of the N-terminus increases the correlation times, resulting in a reduction of signal intensity.
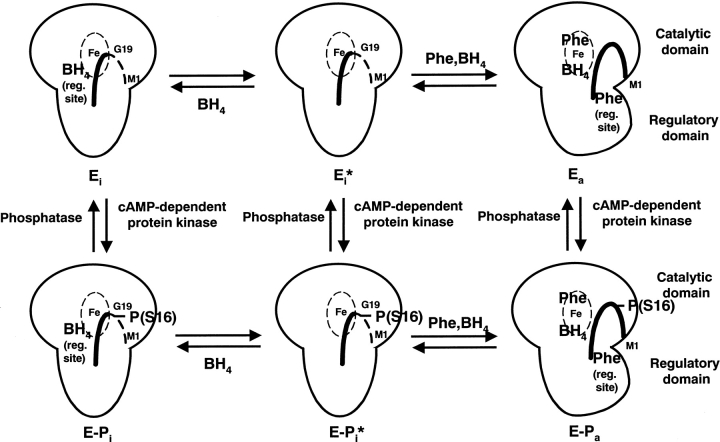
Both the crystallographic results (Kobe et al. 1999) and the NMR studies reported here suggest that the N-terminal 18 residues containing Ser16 are mobile in absence of phenylalanine, in both phosphorylated and unphosphorylated forms; the structures of the phosphorylated and unphosphorylated enzymes differ only locally at the sequence surrounding Ser16. Importantly, we show here that upon the addition of phenylalanine, the mobility of a portion (residues 10–16, but not residues 1–7) of the N-terminal sequence decreases significantly, presumably through the association with the folded core of the protein. Large conformational changes are known to occur in PAH upon phenylalanine binding (Shiman et al. 1979; Hufton et al. 1995; Fitzpatrick 2000), and it is reasonable that a binding site is created for the N-terminal sequence in the phenylalanine-induced conformation. It is possible that the N-terminal sequence is directly involved in phenylalanine binding. Phosphorylation could stabilize the binding of the N-terminal sequence to the core of PAH, through direct electrostatic interaction (Citron et al. 1994; Kowlessur et al. 1995) and/or by affecting the structural properties of the local sequence surrounding the phosphorylation site. Our working model for the regulation of PAH enzyme activity by phosphorylation is schematically shown in Figure 4 ▶.
Fig. 4.
Schematic diagram of the regulation of PAH by phenylalanine, BH4, and phosphorylation. The large object represents a monomer of PAH, with the large protrusion as the catalytic domain and the small protrusion as the regulatory domain. The dashed ellipse with Fe is the active site, and the thick curved line is the N-terminal autoregulatory sequence. The dashed line represents mobile regions, and the solid line represents ordered regions. Little is currently known about the locations of phenylalanine and BH4 binding sites; they are shown in arbitrary locations in the model. The right column represents active forms of PAH, and the left column autoinhibited forms of PAH. Phosphorylation (bottom row) facilitates the phenylalanine-induced conversion from autoinhibited to active form.
Although phosphorylation is the major mechanism of cellular regulation, its consequences have been structurally elucidated in very few systems (Johnson and O'Reilly 1996). In PAH, phosphorylation only has an effect concurrently with the binding of another effector, phenylalanine. Although other examples of concerted action of regulatory events exist, for example, the combination of cyclin binding and phosphorylation in cyclin-dependent protein kinase CDK2 (Pavletich 1999), and dual phosphorylation in MAP kinase ERK2 (Canagarajah et al. 1997), the lack of nonlocalized structural consequences of phosphorylation in PAH in absence of phenylalanine is rather unique. In yeast glycogen phosphorylase, where an N-terminal regulatory sequence similarly blocks the active site of the adjacent monomer in the unphosphorylated form, phosphorylation at Thr10 within this sequence is able on its own to cause movement from the active site (Lin et al. 1997).
Materials and methods
Protein expression, purification, and phosphorylation
PAH1–428, corresponding to residues 1–428 of rat PAH (lacking residues 429–452), was expressed in insect cells and purified using phenyl-Sepharose and DE-52 cellulose chromatography (Kobe et al. 1997). PAH1–428 has been shown to have similar catalytic properties to the full-length PAH and to be similarly activated by phenylalanine (Hufton et al. 1998) and phosphorylation (Kobe et al. 1997), but has a defined dimeric structure, in contrast to full-length PAH, which is a mixture of dimers and tetramers (Hufton et al. 1998). The purified PAH1–428 was phosphorylated using the purified catalytic subunit of cAMP-dependent protein kinase or dephosphorylated using alkaline phosphatase (Boehringer-Mannheim) and then repurified using anion-exchange chromatography on a DE-52 cellulose column (Kobe et al. 1997). The phosphorylation state was assessed by electrospray mass spectrometry (Kobe et al. 1997).
PAH30–428, corresponding to residues 30–428 of rat PAH was expressed in Escherichia coli and purified by ammonium sulfate fractionation, ion exchange chromatography on DE-52 (Whatman) cellulose, and affinity chromatography on a Biogel HTP hydroxyapatite (Biorad) (Jennings et al. 2001).
Peptide synthesis and purification
The peptide corresponding to amino acids 2–20 of rat PAH (AAVVLENGVLSRKLSDFGQ) (PAH2–20) was synthesized using the Applied Biosystems 433A peptide synthesizer, purified by cation exchange chromatography followed by reverse-phase chromatography, and analyzed by quantitative amino acid analysis using a Beckman 6300 amino acid analyzer and electrospray mass spectrometry (Sciex API 111, Perkin-Elmer) (Michell et al. 1996).
PAH2–20 was phosphorylated as described above for PAH1–428, with the peptide purified through a Sepak cartridge followed by drying under vacuum. Analysis of the phosphorylated peptide by MALDI/TOF mass spectrometry confirmed that the peptide was fully phosphorylated on a single residue.
NMR spectroscopy
Immediately prior to NMR measurements, the proteins were concentrated to at least 5 mg/mL (>0.1 mM) in a final volume of 0.5 mL using a Centricon-30, followed by dialysis into 50 mM KH2PO4/K2HPO4 (pH 6.5), containing 50 mM KCl. Phenylalanine activation was carried out in the NMR tube by the addition of solid phenylalanine to a concentration of 200 mM (to push the equilibrium as far as possible towards the activated form). The addition of phenylalanine resulted in no apparent change in pH. Both the unphosphorylated and phosphorylated forms of the PAH2–20 peptide were dissolved in 50 mM KH2PO4/K2HPO4 (pH 6.5) containing 50 mM KCl at a concentration of 1.5 mM for NMR experiments.
Clean two-dimensional (2D) total correlated (TOCSY) spectra (Cavanagh and Rance 1992) were acquired for both protein and peptide samples using 65-msec mixing times. 2D nuclear Overhauser effect (NOESY) spectra of the peptide were acquired with mixing times of 200 to 400 msec. Water signal was suppressed using WATERGATE methods (Liu et al. 1997). Data were collected at 25°C with spectral widths of 7500 Hz in both dimensions and as matrices of 2048 × 256 complex points. The field strength was 600 MHz. Spectra were processed using NMRPipe (Delaglio et al. 1995), typically with cosine bell apodization and zero-filling prior to Fourier transformation followed by base-plane correction, and analyzed in XEASY (Bartels et al. 1995).
Acknowledgments
We thank Frosa Katsis and Bruce Kemp for peptide synthesis. This work was supported by the Australian Research Council (B.K. and P.R.G.) and the Wellcome Trust (B.K.). B.K. is a Wellcome Senior Research Fellow in Medical Science in Australia.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.4560102.
References
- Bartels, C., Xia, T., Billeter, M., Guntert, P., and Wuthrich, K. 1995. The program XEASY for computer-supported NMR spectral analysis of biomolecules. J. Biomol. NMR 6 1–10. [DOI] [PubMed] [Google Scholar]
- Canagarajah, B.J., Khokhlatchev, A., Cobb, M.H., and Goldsmith, E.J. 1997. Activation mechanism of the MAP kinase ERK2 by dual phosphorylation. Cell 90 859–869. [DOI] [PubMed] [Google Scholar]
- Cavanagh, J. and Rance, M. 1992. Suppression of cross-relaxation effects in TOCSY spectra via a modified DIPSI-2 mixing sequence. J. Magn. Reson. 96 670–678. [Google Scholar]
- Citron, B.A., Davis, D.M., and Kaufman, S. 1994. Electrostatic activation of rat phenylalanine hydroxylase. Biochem. Biophys. Res. Commun. 198 174–180. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., and Bax, A. 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6 277–293. [DOI] [PubMed] [Google Scholar]
- Donlon, J. and Kaufman, S. 1978. Glucagon stimulation of rat hepatic phenylalanine hydroxylase through phosphorylation in vivo. J. Biol. Chem. 253 6657–6659. [PubMed] [Google Scholar]
- Doskeland, A.P., Doskeland, S.O., Ogreid, D., and Flatmark, T. 1984. The effect of ligands of phenylalanine 4-monooxygenase on the cAMP-dependent phosphorylation of the enzyme. J. Biol. Chem. 259 11242–11248. [PubMed] [Google Scholar]
- Fitzpatrick, P.F. 2000. The aromatic amino acid hydroxylases. Adv. Enzymol. Relat. Areas Mol. Biol. 74 235–294. [DOI] [PubMed] [Google Scholar]
- Flatmark, T. and Stevens, R.C. 1999. Structural insight into the aromatic amino acid hydroxylases and their disease-related mutant forms. Chem. Rev. 99 2137–2160. [DOI] [PubMed] [Google Scholar]
- Hufton, S.E., Jennings, I.G., and Cotton, R.G.H. 1995. Structure and function of the aromatic amino acid hydroxylases. Biochem. J. 311 353–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1998. Structure/function analysis of the domains required for the multimerisation of phenylalanine hydroxylase. Biochim. Biophys. Acta 1382 295–304. [DOI] [PubMed] [Google Scholar]
- Jennings, I.G., Teh, T., and Kobe, B. 2001. Essential role of the N-terminal autoregulatory sequence in the regulation of phenylalanine hydroxylase. FEBS Lett. 488 196–200. [DOI] [PubMed] [Google Scholar]
- Johnson, L.N. and O'Reilly, M. 1996. Control by phosphorylation. Curr. Opin. Struct. Biol. 6 762–769. [DOI] [PubMed] [Google Scholar]
- Kappock, T.J. and Caradonna, J.P. 1996. Pterin-dependent amino acid hydroxylases. Chem. Rev. 96 2659–2756. [DOI] [PubMed] [Google Scholar]
- Kaufman, S. 1993. The phenylalanine hydroxylating system. Adv. Enzymol. 67 77–264. [DOI] [PubMed] [Google Scholar]
- Kobe, B. and Kemp, B.E. 1999. Active site-directed protein regulation. Nature 402 373–376. [DOI] [PubMed] [Google Scholar]
- Kobe, B., Jennings, I.G., House, C.M., Feil, S.C., Michell, B.J., Tiganis, T., Parker, M.W., Cotton, R.G.H., and Kemp, B.E. 1997. Regulation and crystallization of phosphorylated and dephosphorylated forms of truncated dimeric phenylalanine hydroxylase. Protein Sci. 6 1352–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe, B., Jennings, I.G., House, C.M., Michell, B.J., Goodwill, K.E., Santarsiero, B.D., Stevens, R.C., Cotton, R.G.H., and Kemp, B.E. 1999. Structural basis of autoregulation of phenylalanine hydroxylase. Nat. Struct. Biol. 6 442–448. [DOI] [PubMed] [Google Scholar]
- Kowlessur, D., Yang, X.-J., and Kaufman, S. 1995. Further studies of the role of Ser-16 in the regulation of the activity of phenylalanine hydroxylase. Proc. Natl. Acad. Sci. 92 4743–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry, S.J., Taher, A., Georgopoulos, C., and van der Vies, S.M. 1996. Interplay of structure and disorder in cochaperonin mobile loops. Proc. Natl. Acad. Sci. 93 11622–11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry, S.J., Zeilstra-Ryalls, J., Fayet, O., Georgopoulos, C., and Gierasch, L.M. 1993. Characterization of a functionally important mobile domain of GroES. Nature 364 255–258. [DOI] [PubMed] [Google Scholar]
- Lin, K., Hwang, P., and Fletterick, R.J. 1997. Distinct phosphorylation signals converge at the catalytic center in glycogen phosphorylase. Structure 5 1511–1523. [DOI] [PubMed] [Google Scholar]
- Liu, M., Mao, X.A., Ye, C., Huang, H., Nicholson, J.K., and Lindon, J.C. 1997. Improved WATERGATE pulse sequences for solvent suppression in NMR spectroscopy. J. Magn. Reson. 132 125–129. [Google Scholar]
- Michell, B.J., Stapleton, D., Mitchelhill, K.I., House, C.M., Katsis, F., Witters, L.A., and Kemp, B.E. 1996. Isoform-specific purification and substrate specificity of the 5`-AMP-activated protein kinase. J. Biol. Chem. 271 28445–28450. [DOI] [PubMed] [Google Scholar]
- Nowacki, P., Byck, S., Prevost, L., and Scriver, C.R. 1997. The PAH mutation analysis consortium database: Update 1996. Nucleic Acids Res. 25 139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich, N.P. 1999. Mechanisms of cyclin-dependent kinase regulation: Structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J. Mol. Biol. 287 821–828. [DOI] [PubMed] [Google Scholar]
- Shiman, R. and Gray, D.W. 1980. Substrate activation of phenylalanine hydroxylase. A kinetic characterization. J. Biol. Chem. 255 4793–4800. [PubMed] [Google Scholar]
- Shiman, R., Gray, D.W., and Pater, A. 1979. A simple purification of phenylalanine hydroxylase by substrate-induced hydrophobic chromatography. J. Biol. Chem. 254 11300–11306. [PubMed] [Google Scholar]
- Shiman, R., Mortimore, G.E., Schworer, C.M., and Gray, D.W. 1982. Regulation of phenylalanine hydroxylase activity by phenylalanine in vivo, in vitro, and in perfused rat liver. J. Biol. Chem. 257 11213–11216. [PubMed] [Google Scholar]
- Teigen, K., Froystein, N.Å., and Martinez, A. 1999. The structural basis of the recognition of phenylalanine and pterin cofactors by phenylalanine hydroxylase. Implications for the catalytic mechanism. J. Mol. Biol. 294 807–823. [DOI] [PubMed] [Google Scholar]
- Wang, G.A., Gu, P., and Kaufman, S. 2001. Mutagenesis of the regulatory domain of phenylalanine hydroxylase. Proc. Natl. Acad. Sci. 98 1537–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wretborn, M., Humble, E., Ragnarsson, U., and Engstrom, L. 1980. Amino acid sequence at the phosphorylation site of rat liver phenylalanine hydroxylase and phosphorylation of a corresponding peptide. Biochem. Biophys. Res. Commun. 93 403–408. [DOI] [PubMed] [Google Scholar]