Table 1.
Asp box motifs detected by rigid body searching
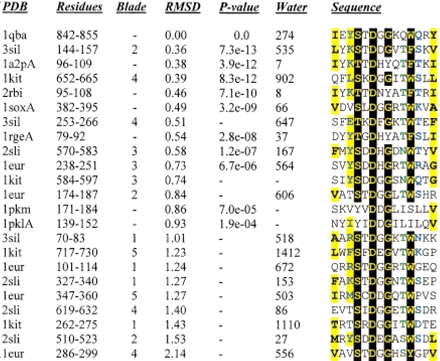 |
The sequences are colored according to Figure 2 ▶. The identifying number of the water molecule associated with each hairpin is given. Two pyruvate kinase structures were detected at values just lower than the 3 × 10−4P-value threshold, and they are not considered by this analysis as containing bona fide Asp boxes due to the absence of a water molecule at positions equivalent to those seen for the sialidases, chitobiase, and ribonucleases (see text for details).
Key to PDB identifiers: 3sil Salmonella typhimurium sialdase; 1a2p Barnase, B. amyloliquefaciens ribonuclease; 1kit Vibrio choerae neuraminidase; 2rbi Mutated binase, B. intermedius ribonuclease; 1sox Gallus gallus sulfite oxidase; 1rge Streptomyces aureofaciens ribonuclease; 2sli Leech trans-sialidase; 1eur Micromonospora viridifaciens sialidase; 1pkm Feline pyruvate kinase; 1pkl Leishmania pyruvate kinase. 1sox & 1qba adopt IG-like folds. 1a2p, 1rge and 2rbi adopt microbial ribonuclease folds. The remaining folds are 6 bladed β propellers. The internal repeat corresponding to each motif is given in the "blade" column for these folds.
