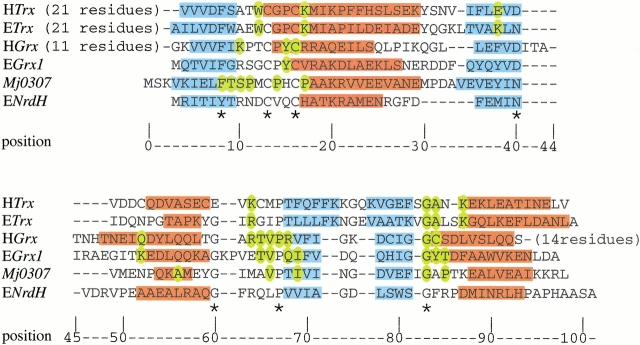
Fig. 8.
Structure-based sequence alignment for H. sapiens Trx (HTrx), E. coli Trx (ETrx), H. sapiens Grx (HGrx), E. coli Grx-1 (EGrx1), Mj0307, and E. coli NrdH (ENrdH). (blue) Residues that adopt a β strand conformation; (red) helical residues. The residues that are important for protein–protein interactions in Trx (Eklund et al. 1991), glutathoine binding in HGrx, and EGrx1 are colored in yellow (Bushweller et al. 1994; Yang et al. 1998). The residues that are conserved in Mj0307, Mt0807, and Af2145 are also colored yellow in the Mj0307 sequence. The residues that are conserved in all of the aligned sequences are indicated by an asterisk below the alignment. The structures for all sequences are accessible in the Protein Databank, except ENrdH, which has its secondary structure derived from the PHD structure prediction (Rost 1996) and sequence homology.