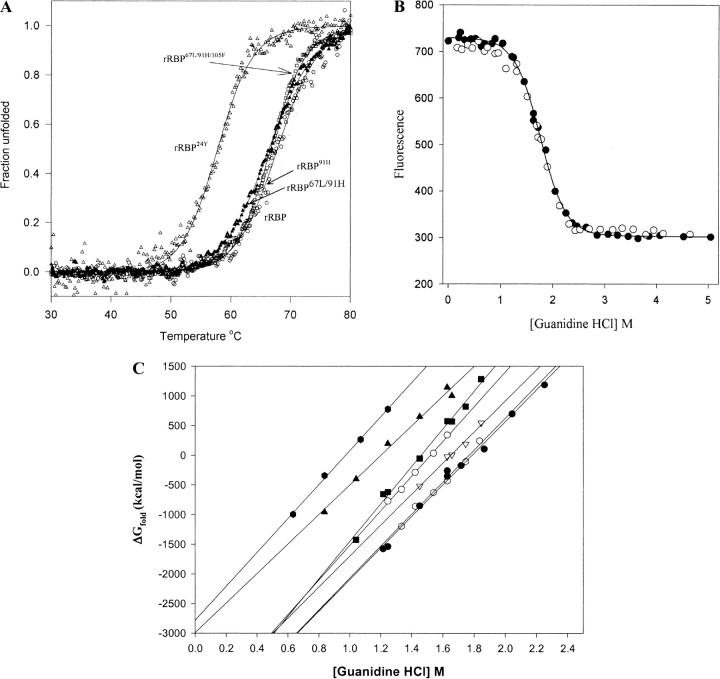
Fig. 7.
Stability studies of the recombinant retinol-binding protein (rRBP) and variants. (A) Thermal unfolding for rRBP and mutants measured by CD ellipticity at 250 nm. All proteins were dissolved in 5 mM phosphate buffer (pH 7.4). Protein samples were heated from 20°C to 80°C at a rate of 50°C/h. The pathlength was 1.0 cm. (B) Solvent-induced equilibrium denaturation and refolding of rRBP monitored by tryptophan fluorescence: (closed circles) Unfolding of rRBP; (open circles) refolding. Protein samples were incubated in 5 mM NaPO4 buffer and a range of GndHCl concentrations (0 to 6 M) for 24 h at 23°C. Fluorescence emission was measured from 329 to 331 nm using an excitation wavelength of 290 nm. Each spectrum is an average of nine scans. (C) Plot of ΔG versus GndHCl concentration for rRBP and mutants. Chemical unfolding was conducted with varying concentrations of GndHCl: rRBP (circles), rRBP24Y (triangles), rRBP22A/24F (hexagon), rRBP67L/91H/105F (squares), rRBP91H (open hexagons), rRBP105F (open circles), and rRBP67L/91H (open triangles).