Fig. 3.
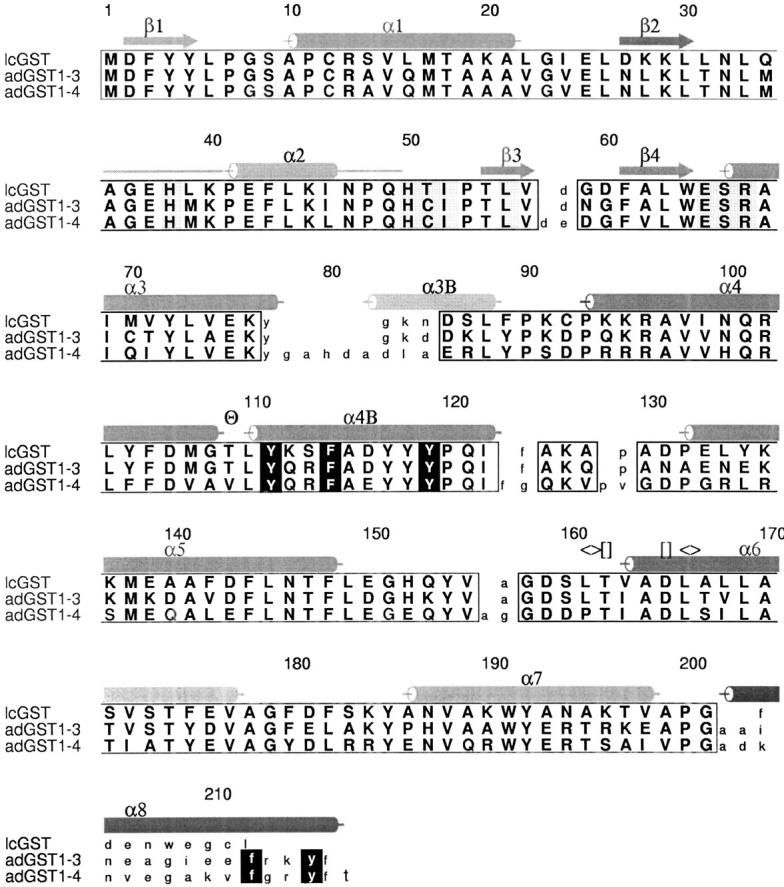
Structure-based sequence alignment of insect GSTs. Secondary structure is indicated above the sequences in red. The kink in helix α4 is indicated (Θ). The hydrophobic staple at the base of helix α6 is indicated (<>), as is the N-terminal capping m ([]). Regions of high structural conservation are boxed with amino acids indicated in bold upper case. Residue numbers are those for AdGST1–4. The region of AdGST1–4 with no electron density (around helix α2) is indicated with a horizontal line. Helix α8, unique to the AdGST isozymes is indicated. The helix observed linking the N- and C-terminal domains of AdGST1–4 is indicated as α3B. G-site residues have shaded backgrounds. H-site residues have white letters on a black background.
