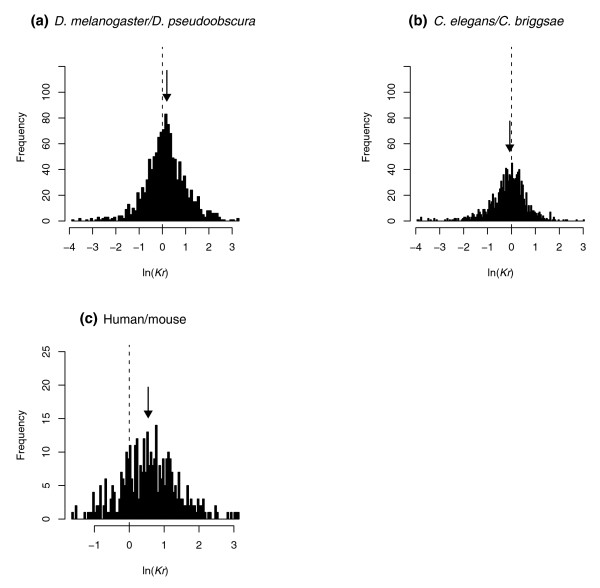
Figure 6.
Exon cores and flanks evolve at different rates. Histograms of logged Kr ratios (KAc/KAf), using 100 bins, for (a) D. melanogaster-D. pseudoobscura orthologous genes (N = 1,237), (b) C. elegans-C. briggsae orthologous genes (N = 1,102), and (c) human-mouse orthologous genes (N = 360) with a minimum of 300 bp of concatenated middle and flanking sequence of exons are plotted. The dashed line in each graph indicates ln(Kr) = 0, the point at which middle and flanking sections evolve at the same average rate. The arrows indicate the median logged Kr ratios of (a) 0.128, (b) -0.065, and (c) 0.559, respectively. All three are significantly different from the null expectation of ln(Kr) = 0 (P < 0.0001). Note the much more marked departure from the null expectation in the mammalian dataset.