Short abstract
The sequencing of the genome of a female rhesus macaque (Macaca mulatta) of Indian origin will provide us with biomedical and evolutionary insights into both humans and Old World monkeys.
Abstract
The sequencing of the genome of a female rhesus macaque (Macaca mulatta) of Indian origin will provide us with biomedical and evolutionary insights into both humans and Old World monkeys.
The recently published draft of the rhesus macaque (Macaca mulatta) genome from the Rhesus Macaque Genome Sequencing and Analysis Consortium [1] follows that of the chimpanzee [2] by only a year and a half, and now gives us three primate genome sequences, including our own. Of the 2.87 gigabases sequenced at 5.2-fold coverage, and containing approximately 20,000 predicted genetic loci, regions of the macaque genome that could be aligned to the human genome sequence were 93.5% identical (90.76% when small insertions and deletions are included). Compared with the human-chimpanzee difference of 98.77%, which sometimes gives sequences that are too similar to draw meaningful comparisons, and the human-mouse difference of 69.1%, which gives sequences often too divergent to be useful, the macaque sequences provide Goldilocks' 'just right' for many types of analyses.
The macaque and human evolution
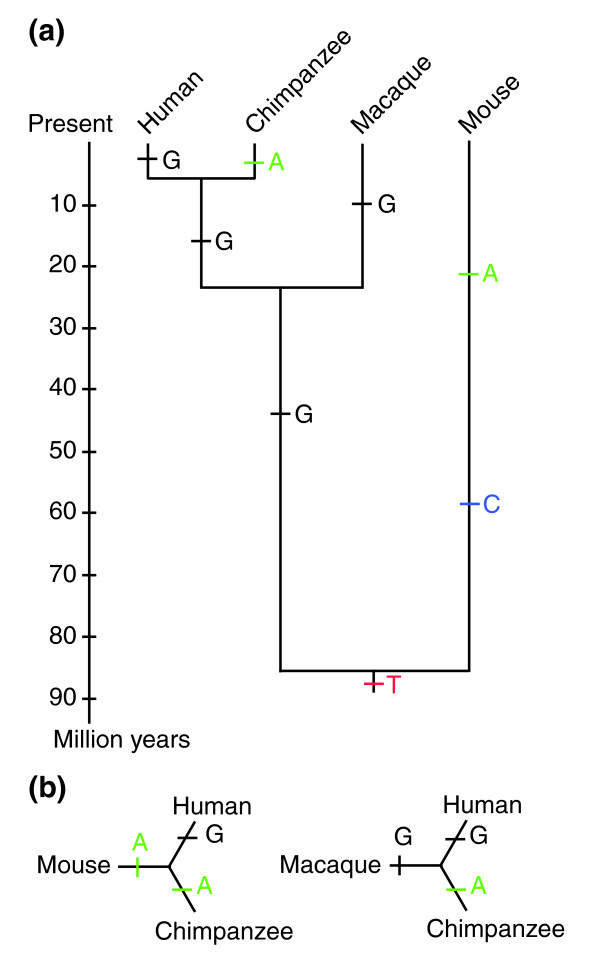
One of the hopes and justifications for sequencing the chimpanzee genome was that it would allow us to identify the genetic changes 'that make us human'. Once chimpanzee genome sequences started to become available, papers quickly appeared, searching for unique genetic changes along the human lineage after we separated from chimpanzees. In the absence of other primate genome sequences, the mouse was used for comparison with chimpanzee and human [3]. However, given the relatively deep evolutionary divergence of the mouse and primate lineages, of the order of at least 70 million years ago, so many changes could have occurred either along the mouse lineage or on the long branch leading to the common ancestor of humans and chimpanzees that we cannot with much confidence estimate what nucleotide was present in any position in that ancestor. Thus, we were not able to reasonably estimate whether a given difference between the chimp and human genomes had occurred in the human lineage or in the chimpanzee lineage (Figure 1). Using the macaque genome as a comparison, however, we can now place changes on a lineage far more reliably, because the probability of convergent changes is much smaller than with the mouse.
Figure 1.
The macaque is a better outgroup than the mouse for inferring the history of sequence changes in human and chimpanzee genomes. (a) The scaled phylogeny of primates with respect to the mouse. Over long evolutionary periods, multiple mutations are likely to occur at the same position in the genome, obscuring that base's true evolutionary history. This is indicated here by the change of the initial T to a C and later to an A in the mouse genome, and the change from the T to a G in the primate line, and later to an A in the chimpanzee line only. (b) If a distantly related species (the mouse) is used as the outgroup in a comparison of the human and chimpanzee genomes, this can lead to the mistaken conclusion that a unique mutation has occurred along the human lineage, as demonstrated in the diagram on the left. When the genomes are compared using a more closely related outgroup (the macaque) the more probable history of this difference is revealed, as shown in the diagram on the right.
Screens for positively selected changes between chimpanzees and humans using the mouse genome as an outgroup initially suggested that selected changes were more numerous in the human lineage than in the chimp lineage [3]. Other studies found a possibly accelerated rate of change in conserved noncoding regions in the human lineage [4]. These observations were readily accepted, in part because they supported our naturally anthropocentric view that humans are special and so there should be a molecular signature of our uniqueness. More recent analyses using the more closely related macaque as the outgroup suggest, however, that a greater number of positively selected changes has in fact occurred along the chimpanzee lineage, leaving humans as the more 'primitive' species from a genomic standpoint [5,6]. This is somewhat surprising, given that overall the skeletons of our 5-6-million-year-old ancestors look remarkably chimpanzee-like. With respect to our extremely large and complex brain, studies using the mouse as outgroup proposed an accelerated rate of evolution in nervous-system genes in humans [7]. But, perhaps no longer surprisingly, a recent study using the macaque genome for comparison showed that even genes expressed specifically in the brain were found to be under no greater selection in humans than in chimpanzees [8]. Thus, we still do not know the molecular basis for the evolution of the uniquely large human brain.
The macaque genome has also benefited our understanding of the human genome in other ways. For example, the method called 'phylogenetic shadowing' involves the comparison of DNA sequences across multiple species to reveal conservative sequence blocks. Such conserved regions may be putative exons, regulatory elements, or otherwise functionally significant [9,10]. By comparing sequences of closely related species (for example, between primates, rather than distinctly related animals), the rare changes within these 'least variable' regions may highlight the critical mutations that make a species unique.
Macaque diversity and its implications for biomedical research
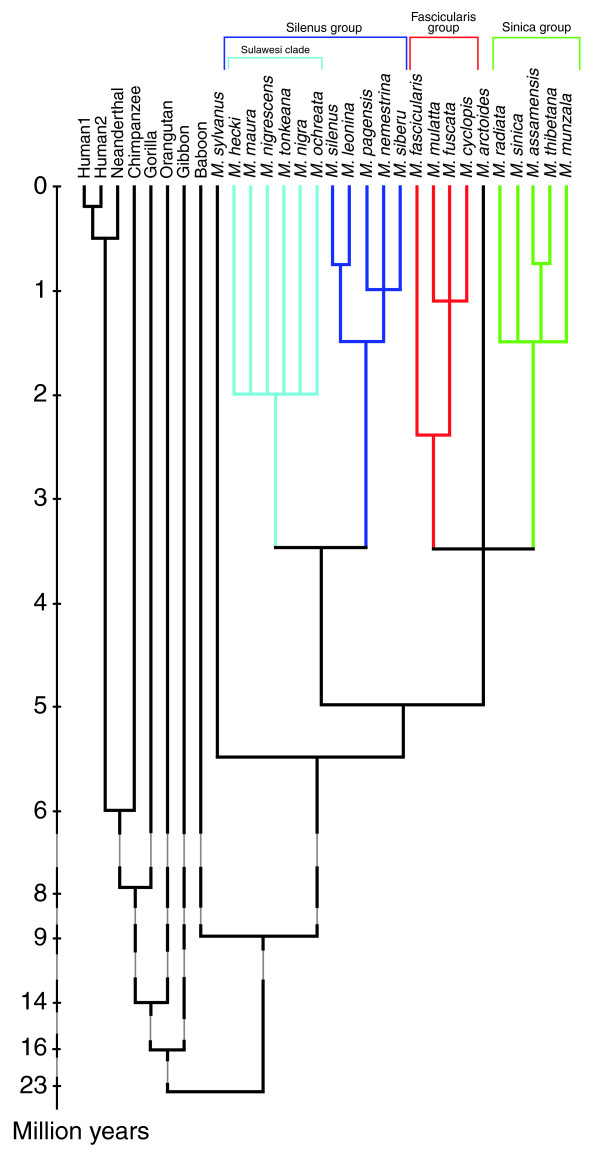
The macaque genome not only opens up whole new areas of understanding of an important model organism and provides us with an important perspective on human evolution, it also gives us more tools for studying the incredibly diverse array of interesting monkeys that are the most widely used primates in medical research; as well as M. mulatta, these include the long-tailed macaque (M. fascicularis), the Japanese macaque (M. fuscata), the pig-tailed macaque (M. nemestrina) and the bonnet macaque (M. radiata). While it is commonly stated that macaques last shared a common ancestor with humans, chimpanzees and other apes around 25 million years ago, both the fossil record and recent molecular analyses suggest a slightly more recent date, in the order of 23 million years ago (Figure 2). The macaque lineage itself originated approximately 9 million years ago, most probably in Africa, with the diversification of living macaques beginning around 5-6 million years ago [11].
Figure 2.
Consensus phylogeny of the genus Macaca placed within the evolutionary history of several Old World primate lineages. Except for the Barbary macaque (M. sylvanus) found in North Africa and Gibraltar, and the stump-tailed or bear macaque (M. arctoides) found in the border regions of India, China and Malaysia, macaques are divided into three main species groups. Divergence patterns and times within Macaca are taken from [11], while those among outgroup lineages are taken from [16]. Note the deep divergence times among the macaques. The dates of the oldest bifurcations are comparable to that estimated for the human-chimpanzee split, and even the youngest bifurcations pre-date the origin of anatomically modern humans by several hundred thousand years. Individual macaque species are likely to have accrued significant genetic diversity, and researchers need to take this into account when designing and interpreting the results of biomedical tests using these animals.
It is important to recognize the relatively deep evolutionary history of macaques when planning or interpreting biomedical studies. The five species mentioned above make up the majority of macaques used in such studies, and diverged from each other up to 5 million years ago. This means that as far as evolutionary divergence goes, substituting one species for another is akin to substituting humans for chimps. It is therefore highly desirable that the genetic backgrounds of the macaque species used in research, and their differences from each other, should be fully assessed.
It is already known that different species and subspecies of macaques react differently and show different levels of pathogenesis with respect to two of the most widely studied human infectious diseases, AIDS and malaria. For instance, rhesus macaques of Indian origin (like the one whose genome was sequenced [2]) progress much more rapidly to simian AIDS upon infection with simian immunodeficiency virus (SIV) compared with rhesus macaques of Chinese origin [12,13]. Rhesus macaques and mainland Malaysian populations of long-tailed macaques show greater susceptibility and pathogenesis to some strains of malaria than do long-tailed macaques from the Philippines, which are far less susceptible and do not get as sick [14]. This correlates with the observation that mainland populations of long-tailed macaques are known to hybridize with rhesus macaques, as demonstrated by the lengths of their tails and molecular markers.
To investigate the diversity of these monkeys, nearly 1,500 single-nucleotide polymorphisms (SNPs) were typed in nine Chinese and 38 Indian rhesus macaques as part of the rhesus macaque genome project [15]. The monkeys of Indian origin show much less variability than the Chinese animals, which again needs to be taken into account in any studies using macaques. Another interesting finding from the rhesus genome project is that a significant number of disease-causing or disease-associated alleles in humans are found in macaques or in the inferred common ancestor of chimpanzees and humans (when using the rhesus macaque as the outgroup). Since these alleles do not cause disease in macaques, we must be more cautious in using non-human primate models (and, obviously, non-primate models) in investigating human genetic diseases.
What is also clear from the studies so far is the importance of finished rather than draft genomes for detailed comparative analyses. Accurate sequences, and thus alignments, are critical when assessing polymorphism, the influence of selection and ancestral states. With a variety of other primate genomes currently being sequenced or in the planning stages - including gorilla, orangutan, gibbon, marmoset, tarsier, galago and mouse lemur - breakthroughs in understanding the evolutionary history and biology we share with our closest living relatives will continue to occur at an increasing pace.
Acknowledgments
Acknowledgements
We would like to thank Christina Bergey for assistance with the figures and Eleni Nikitopoulos for her comments on the manuscript.
References
- Rhesus Macaque Genome Sequencing and Analysis Consortium Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Clark AG, Glanowski S, Nielsen R, Thomas PD, Kejariwal A, Todd MA, Tanenbaum DM, Civello D, Lu F, Murphy B, et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–1963. doi: 10.1126/science.1088821. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Noonan JP, Pääbo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- Bakewell MA, Shi P, Zhang J. More genes underwent positive selection in chimpanzee evolution than in human evolution. Proc Natl Acad Sci USA. 2007;104:7489–7494. doi: 10.1073/pnas.0701705104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X-J, Zheng H-K, Wang J, Wwang W, Su B. Detecting lineage-specific adaptive evolution of brain-expressed genes in human using rhesus macaque as outgroup. Genomics. 2006;88:745–751. doi: 10.1016/j.ygeno.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Dorus S, Vallender EJ, Evans PD, Anderson JR, Gilbert SL, Mahowald M, Wyckoff GJ, Malcom CM, Lahn BT. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. [DOI] [PubMed] [Google Scholar]
- Shi P, Bakewell MA, Zhang J. Did brain-specific genes evolve faster in humans than in chimpanzees. Trends Genet. 2006;22:608–613. doi: 10.1016/j.tig.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Boffelli D, McAuliffe J, Ovcharenko D, Lewis KD, Ovharenko I, Pachter L, Rubin EM. Phylogenetic shadowing of primate sequences to find functional regions of the human genome. Science. 2003;299:1391–1394. doi: 10.1126/science.1081331. [DOI] [PubMed] [Google Scholar]
- Wang Q-F, Prabhakar S, Chanan S, Cheng J-F, Rubin EM, Boffelli D. Detection of weakly conserved ancestral mammalian regulatory sequences in primate comparisons. Genome Biol. 2007;8:R1. doi: 10.1186/gb-2007-8-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi AJ, Morales JC, Melnick DJ. Paternal, maternal, and biparental molecular markers provide unique windows onto the evolutionary history of macaque monkeys. Evolution. 2003;57:1419–1435. doi: 10.1111/j.0014-3820.2003.tb00349.x. [DOI] [PubMed] [Google Scholar]
- Ling B, Veazey RS, Luckay A, Penedo C, Xu K, Lifson JD, Marx PA. SIV(mac) pathogensis in rhesus macaques of Chinese and Indian origin compared with primary HIV infections in humans. AIDS. 2002;16:1489–1496. doi: 10.1097/00002030-200207260-00005. [DOI] [PubMed] [Google Scholar]
- Trichel AM, Rajakumar PA, Murphey-Corb M. Species-specific variation in SIV disease progression between Chinese and Indian subspecies of rhesus macaque. J Med Primatol. 2002;31:171–178. doi: 10.1034/j.1600-0684.2002.02003.x. [DOI] [PubMed] [Google Scholar]
- Schmidt LH, Fradkin R, Harrison J, Rossan R. Differences in the virulence of Plasmodium knowlesi for Macaca irus (fascicularis) of Philippine and Malayan origins. Am J Trop Med. 1977;26:612–622. doi: 10.4269/ajtmh.1977.26.612. [DOI] [PubMed] [Google Scholar]
- Hernandez RD, Hubisz MJ, Wheeler DA, Smith DG, Ferguson B, Rogers J, Nazareth L, Indap A, Bourquin T, McPherson J, et al. Demographic histories and patterns of linkage disequilibrium in Chinese and Indian rhesus macaques. Science. 2007;316:240–243. doi: 10.1126/science.1140462. [DOI] [PubMed] [Google Scholar]
- Springer MS, Murphy WJ, Eizirik E, O'Brien SJ. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc Natl Acad Sci USA. 2003;100:1056–1061. doi: 10.1073/pnas.0334222100. [DOI] [PMC free article] [PubMed] [Google Scholar]