Figure 2.
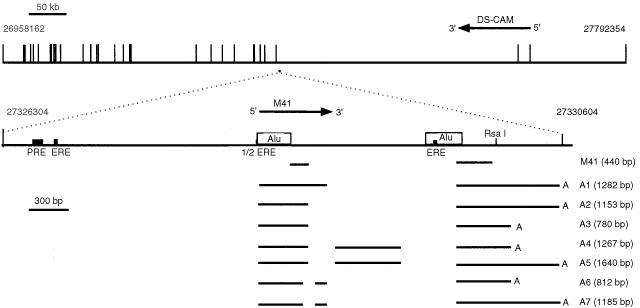
Alignment of the M41 gene and M41 transcripts. The gene region of chromosome 21q22.1 is shown at the top with the exons of the DS-CAM gene shown as vertical bars. The location of the M41 gene is shown as a square to scale beneath the genomic DNA and expanded beneath. The location of response elements for oestrogen (ERE) and progesterone (PRE) and Alu sequences are shown. The region of the original M41 cDNA isolated from the MCF-7 subtracted library, and processing variants of M41 mRNA (A1–A7) obtained from the RACE reactions are shown as horizontal bars with gaps indicating the intronic regions. Numbering refers to the base number of contig NT_011512.3. The 5′ intron exon boundaries revealed by the RACE reactions at 27328651, 27328698, 27328827, 27329381, and the 3′ intron exon boundaries at 27328749, 27328894, 27329821 (enumeration of contig NT_011512.3) all contained conserved GT / AG sequences and exhibited 62.5, 75, 87.5, 100, 75, 93.75, and 81.25% identity, respectively, to the consensus sequences for 5′ or 3′ intron exon boundaries (Mount, 1982). The symbols (A) at the ends of the lines indicate the poly(A) addition sites. Variants contained one of two alternative poly(A) addition sites (at 27330601 and 27330228 of NT_011512.3), with a consensus AATAAA poly(A)-addition signal, 29 and 19 nucleotides upstream of the poly(A)-addition sites, respectively. The lengths of the proposed mRNA variants arising from the 7 alternatively-spliced exons correspond broadly to the sizes of two of the bands observed in the Northern blot for the mammary tumour cell line MCF-7 (Figure 1), strongly suggesting that the RACE products are defining near full length mRNAs. Horizontal arrows indicate the direction of transcription of the genes.
