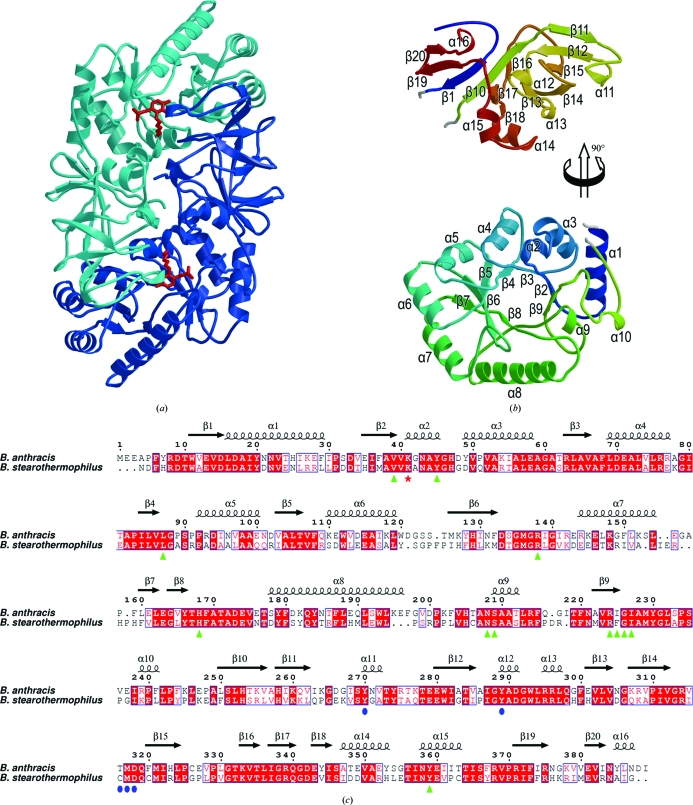
Figure 3.
The structure of B. anthracis alanine racemase. (a) Overall structure of the alanine racemase dimer showing the subunits in light and dark blue and indicating the positions of the active sites by red sticks for the Lys41–PLP adduct. (b) Secondary structure of the monomer (coloured from blue at the N-terminus to red at the C-terminus). For clarity, the C-terminal domain (top) is moved up and rotated 90° about a vertical axis with respect to the N-terminal domain (bottom). Secondary-structural elements are labelled. (c) A sequence alignment based on structure (calculated using SHP) between the alanine racemases of B. anthracis and B. stearothermophilus (PDB code 1sft; Shaw et al., 1997 ▶). The residue numbering and secondary-structural assignments refer to the B. anthracis sequence. Lys41, which forms aSchiff base with PLP, is indicated by a red asterisk. Residues in the B. anthracis structure that interact with PLP (any interatomic distance less than 4.0 Å) are indicated by green triangles, while the additional residues that interact with PLP–l-Ala-P/PLP–d-Ala-P are indicated by blue circles.