Abstract
The p53 tumor suppressor is often disrupted in human cancers by the acquisition of missense mutations. We generated mice with a missense mutation at codon 172 that mimics the p53R175H hot spot mutation in human cancer. p53 homozygous mutant mice have unstable mutant p53 in normal cells and stabilize mutant p53 in some but not all tumors. To investigate the significance of these data, we examined the regulation of mutant p53 stability by Mdm2, an E3 ubiquitin ligase that targets p53 for degradation, and p16INK4a, a member of the Rb tumor suppressor pathway. Mice lacking Mdm2 or p16INK4a stabilized mutant p53, and revealed an earlier age of tumor onset than p53 mutant mice and a gain-of-function metastatic phenotype. Analysis of tumors from p53 homozygous mutant mice with stable p53 revealed defects in the Rb pathway. Additionally, ionizing radiation stabilizes wild-type and mutant p53. Thus, the stabilization of mutant p53 is not a given but it is a prerequisite for its gain-of-function phenotype. Since mutant p53 stability mimics that of wild-type p53, these data indicate that drugs aimed at activating wild-type p53 will also stabilize mutant p53 with dire consequences.
Keywords: Metastasis, mouse models, gain of function, p53 stability
DNA damage, oncogene activation, and hypoxia stabilize wild-type p53 to induce cell cycle arrest, apoptosis, or senescence (Prives and Hall 1999; Vousden 2006; Helton and Chen 2007). Because of these potent activities, the p53 tumor suppressor is a frequent target for mutagenesis in tumor development. The most common p53 alterations are those with missense mutations in the DNA-binding domain rather than gene deletions. Single amino acid changes in p53 that disrupt DNA binding also impair transcriptional activity. Additionally, mutant p53 has an increased half-life and thus accumulates in tumor cells, resulting in the emergence of gain-of-function phenotypes (Dittmer et al. 1993; Sigal and Rotter 2000; Lozano 2007; Weisz et al. 2007).
Various signals regulate wild-type p53 at multiple levels (Giaccia and Kastan 1998; Lavin and Gueven 2006; Horn and Vousden 2007). Regulation is mainly at the protein rather than the transcriptional level and alters p53 stability. p53 stability is mainly mediated by post-translational modifications such as phosphorylation, methylation, sumoylation, ubiquitination, and acetylation (Meek 1994; Gu and Roeder 1997; Sakaguchi et al. 1998; Appella and Anderson 2001; Brooks and Gu 2003; Chuikov et al. 2004). These post-translational modifications occur in response to typical cellular stresses such as DNA damage and oncogene activation. For example, in response to DNA damage, p53 becomes phosphorylated at Ser 15, disrupting its interaction with Mdm2, an E3 ubiquitin ligase that targets p53 for proteasomal degradation (Haupt et al. 1997; Honda et al. 1997; Kubbutat et al. 1997; Shieh et al. 1997; Canman et al. 1998). An alternative mechanism of increasing p53 levels in response to radiation is an increase in translation of p53 mRNA (Takagi et al. 2005). Oncogenic signals such as activating Ras mutations or increased c-Myc expression also stabilize p53 in normal cells and result in senescence or apoptosis, respectively (Hermeking and Eick 1994; Wagner et al. 1994; Serrano et al. 1997; Ferbeyre et al. 2002). Oncogenic activation of p53 is in part mediated via the tumor suppressor p19ARF (Palmero et al. 1998; Sherr 1998; Eischen et al. 1999). In general, p19ARF levels are undetectable in normal tissues, but upon disruption of normal signaling and proliferation, p19ARF levels increase. p19ARF interacts with Mdm2 and this interaction releases inhibition of p53, leading to its activation (Kamijo et al. 1998; Pomerantz et al. 1998; Zhang et al. 1998).
p19ARF is produced from use of an alternative first exon at the INK4a locus (Quelle et al. 1995). This same locus encodes p16INK4a, an inhibitor of the Cyclin-D/Cdk complexes that phosphorylate Rb. Inhibition of Rb phosphorylation induces cell cycle arrest (Medema et al. 1995). Deletion of p16INK4a, overexpression of Cyclin-D, or mutations of Rb contribute to activation of p19ARF and sequestration of Mdm2, and thus activation of p53 (Sherr and McCormick 2002). The impact of these regulatory signals on mutant p53 has not been explored.
Recently, knock-in alleles of murine p53 have been generated in mice to study the functions of mutant p53 in tumorigenesis in vivo. Mice with mutations at amino acids 172 or 270, corresponding to the human hot spot mutations at amino acids 175 and 273, respectively, show gain-of-function phenotypes (Lang et al. 2004; Olive et al. 2004). Heterozygous mice with these mutations exhibit a metastatic phenotype that is rarely observed in p53+/− mice. In tumor cells in culture, mutant p53 binds and inhibits the functions of p63 and p73. Furthermore, p53-null cells mimic the gain-of-function phenotypes of mutant p53 cells upon down-modulation of p63 and p73 (Lang et al. 2004). Importantly, p53+/− mice, upon concomitant deletion of one allele of either p63 or p73, show an increase in metastasis, emphasizing the importance of this mechanism in vivo (Flores et al. 2005). Additional gain-of-function phenotypes include changes in tumor spectrum. Mice with one mutation at amino acid 270 and one null allele show a significant increase in carcinomas (Olive et al. 2004). Moreover, mice homozygous for a murine/human fusion p53 protein with a mutation at amino acid 248 had a greater number of tumor types when compared with p53−/− mice (Song et al. 2007). These changes in metastases and tumor spectrum support the hypothesis that mutant p53 leads to gain-of-function phenotypes in vivo.
Despite these findings, tumor incidence and survival of mice heterozygous or homozygous for a p53 missense mutation did not vary from p53+/− and p53−/− mice, respectively. In this study, we show that mutant p53 is inherently unstable in normal cells and that tumor-specific changes stabilize mutant p53. Mutant p53 is also regulated by Mdm2 in a manner similar to wild-type p53. Accordingly, homozygous mutant mice lacking Mdm2 exhibited increased mutant p53 levels, increased tumor incidence, decreased survival, and a metastatic phenotype when compared with mice homozygous for mutant p53 alone and with p53/Mdm2 double-null mice. Lastly, p16INK4a loss, a common event in human cancers (Kamb et al. 1994; Nobori et al. 1994), also stabilized mutant p53 in vivo and yielded a metastatic phenotype. We also found that radiation stabilized mutant p53. Thus, mutant p53 is regulated like wild-type p53 and its stabilization contributes to its gain-of-function phenotype.
Results
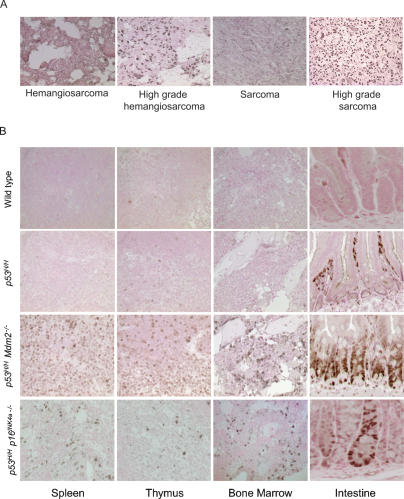
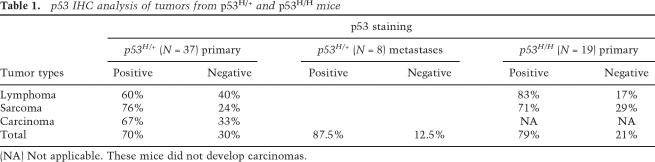
Mutant p53 is often stable in human cancers, and immunohistochemistry (IHC) to detect stable p53 is used as a diagnostic indicator (Hall and Lane 1994). We therefore performed an analysis of mutant p53 levels in tumors from mice carrying the p53515A allele (abbreviated p53H because this mutation changes arginine to histidine at amino acid 172) (see Lang et al. 2004 for details). Homozygous mutant p53H/H mice expressed detectable p53 in 79% of tumors, indicating that 21% of tumors had no detectable mutant p53 (Fig. 1A; Table 1). Analysis of tumors from heterozygous mutant p53H/+ mice indicated that only 70% of these primary tumors had detectable p53 (Table 1). p53 was also not detected in surrounding normal tissues of tumors from p53H/H and p53H/+ mice (Fig. 1A; data not shown). These data suggest that tumor-specific alterations lead to detectable p53 protein levels in most but not all tumors that develop in p53H mutant mice.
Figure 1.
Tumor-specific changes stabilize mutant p53. (A) p53 IHC of hemangiosarcomas and sarcomas from p53H/H mice. Dark nuclear staining denotes high mutant p53 levels. (B) Mutant p53 is inherently unstable in normal tissues but stabilized in the absence of Mdm2 or p16INK4a. p53 IHC of several tissues from wild-type, p53H/H, p53H/H Mdm2−/−, and p53H/H p16INK4a−/− mice. Nonspecific staining in the lumen of the intestine is sometimes observed as in the p53H/H sample.
Table 1.
p53 IHC analysis of tumors from p53H/+ and p53H/H mice
(NA) Not applicable. These mice did not develop carcinomas.
We next characterized p53 stability in normal tissues from p53H/H and p53H/+ mice by IHC. p53 was undetectable by immunostaining in normal tissues examined in homozygous mutant p53H/H mice and in p53H/+ mice, similar to wild-type mice (Fig. 1B; Supplemental Fig. S1; data not shown). Normal tissues examined include lung, spleen, thymus, heart, brain, bone marrow, pancreas, liver, kidney, and intestine. Spleen and thymus tissues showed occasional sporadic staining. These data indicate that the mutant p53R172H protein in vivo in normal tissues is undetectable, similar to wild-type p53 protein.
Mdm2, a negative regulator of p53, is an E3 ubiquitin ligase that targets wild-type p53 for degradation (Haupt et al. 1997; Honda et al. 1997; Kubbutat et al. 1997). Mdm2-null mice are embryonic lethal due to stabilization of p53 and massive apoptosis, a phenotype that is rescued by loss of p53 (Jones et al. 1995; Montes de Oca Luna et al. 1995; Chavez-Reyes et al. 2003). To address the in vivo significance of Mdm2 in regulating the stability of mutant p53, we crossed mice containing the Mdm2-null allele with mice containing the p53H allele. p53H/H Mdm2−/− mice were born at the expected Mendelian ratio and appeared normal, demonstrating the true loss-of-function nature of this missense mutation. Of 88 mice born from p53H/H Mdm2+/− intercrosses, 24 were Mdm2-null, 41 were Mdm2 heterozygous, and 23 were Mdm2 wild type. To assay the levels of mutant p53 in the absence of Mdm2, we performed p53 IHC on normal tissues from double-mutant p53H/H Mdm2−/− mice at 5 wk of age. Mutant p53 was clearly visible in lung, spleen, thymus, heart, brain, bone marrow, pancreas, kidney, intestine, and female genitals (Fig. 1B; Supplemental Fig. S1; data not shown). In some tissues, staining was localized. For example, in the pancreas and kidney, mutant p53 was localized to the islets of Langerhans and the glomerulus, respectively (Supplemental Fig. S1). The only tissue examined lacking detectable mutant p53 was the liver (data not shown). Thus, in p53H/H mice, endogenous levels of Mdm2 regulate the stability of the p53R172H mutant protein in specific cell types of most tissues. The data also suggest that other factors contribute to p53 stability as some, but not all, cells of various tissues stabilized mutant p53.
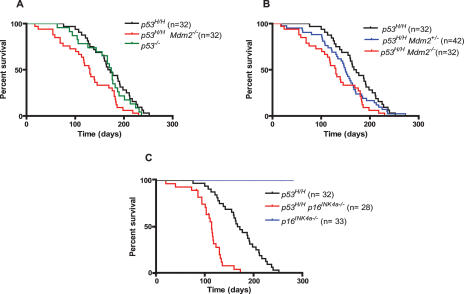
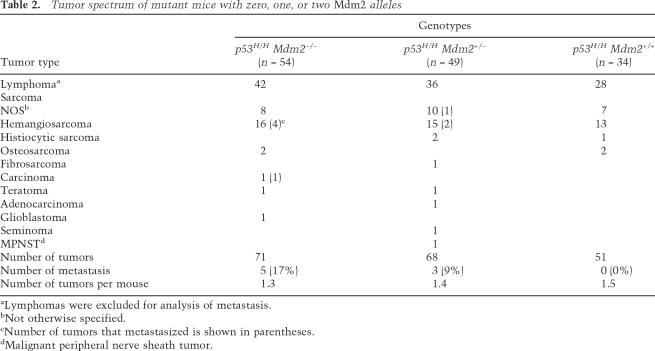
The viability of p53H/H Mdm2−/− mice allowed us to examine tumor incidence and survival. The survival curves of p53−/−, p53−/− Mdm2−/−, and p53H/H control mice are indistinguishable (Fig. 2A; data not shown), as published previously (McDonnell et al. 1999; Lang et al. 2004; Olive et al. 2004). However, p53H/H mice lacking Mdm2 died significantly earlier than p53H/H mice (P = 0.0036) with median survival of 130 and 170 d, respectively (Fig. 2A,B). p53H/H mice heterozygous for Mdm2 had an intermediate survival curve (Fig. 2B). p53H/H mice, regardless of Mdm2 status, developed mostly lymphomas and sarcomas, and no significant differences were observed in the tumor spectrum or in tumor number per mouse between these genotypes (Table 2). Thus, loss of Mdm2 altered tumor onset but not the type of tumors that developed in p53H/H mutant mice.
Figure 2.
Increased tumorigenesis in mutant p53 mice lacking Mdm2 or p16INK4a. (A) Kaplan-Meier survival curves of p53H/H Mdm2−/− compared with p53H/H and p53−/− mice. (B) Kaplan-Meier survival curves of p53H/H mice compared with p53H/H Mdm2−/− and p53H/H Mdm2+/− mice. (C) Kaplan-Meier survival curves of p53H/H p16INK4a−/− mice compared with p53H/H and p16INK4a−/− mice.
Table 2.
Tumor spectrum of mutant mice with zero, one, or two Mdm2 alleles
aLymphomas were excluded for analysis of metastasis.
bNot otherwise specified.
cNumber of tumors that metastasized is shown in parentheses.
dMalignant peripheral nerve sheath tumor.
The one obvious in vivo phenotype of p53H/+ mice missing in p53+/− mice is the metastatic phenotype (Lang et al. 2004; Olive et al. 2004). In contrast, p53H/H mice never developed metastases, perhaps due to the timing of acquisition of other alterations required to stabilize mutant p53R172H (Table 2; Lang et al. 2004). We therefore postulated that stabilization of p53R172H protein contributed to the metastatic phenotype. We monitored the metastatic frequency in p53H/H mice with zero, one, or two null alleles of Mdm2. While p53H/H mice never developed metastases, a dose-dependent increase in the frequency of metastasis occurred upon deletion of one or two alleles of Mdm2. Hence, p53H/H Mdm2+/− mice had a 9% frequency of metastasis, while p53H/H Mdm2−/− mice had a 17% frequency of metastasis (Table 2). Thus, loss of Mdm2 stabilized the p53R172H protein and unveiled a metastatic phenotype in these mutant mice.
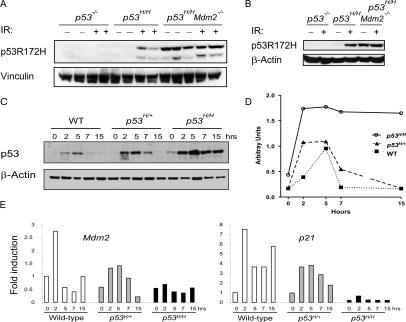
As indicated above, the regulation of p53R172H was similar to that of wild-type p53 in vivo in the absence of Mdm2, suggesting that mechanisms that regulate wild-type p53 regulate mutant p53 as well. Since DNA damage stabilizes wild-type p53 (Kastan et al. 1991), we examined whether DNA damage stabilized mutant p53. These experiments were performed in vivo, as mutant p53 became stable within two to three passages in tissue culture (data not shown). p53H/H and p53H/H Mdm2−/− mice were subjected to 5 Gy whole-body ionizing radiation (IR) and sacrificed 5 h after treatment. p53R172H was stabilized by IR in the spleen and thymus of homozygous mutant mice (Fig. 3A,B). Moreover, deletion of Mdm2 resulted in increased basal levels of p53R172H (as predicted from IHC experiments), which did not further increase after IR (Fig. 3A,B).
Figure 3.
IR stabilized mutant p53 similar to Mdm2 loss in vivo. (A) p53-null, p53H/H, and p53H/H Mdm2−/− mice were irradiated with 5 Gy and sacrificed 5 h after treatment. Spleens were isolated from irradiated and nonirradiated mice, and protein extracts from these tissues were subjected to Western blot analysis with p53 and vinculin (control) antibodies. (B) As in A, irradiated and nonirradiated p53-null, p53H/H, and p53H/H Mdm2−/− mice were sacrificed and protein extracts from thymuses were subjected to p53 and β-actin (control) antibodies. (C) Wild-type, p53H/+, or p53H/H mice were irradiated with 5 Gy and sacrificed at 2, 5, 7, and 15 h after irradiation. Protein extracts were made from spleens of nonirradiated and irradiated mice and subjected to Western blot analysis with p53 and β-actin (control) antibodies. (D) Quantification of p53 protein levels after IR. (E) Wild-type, p53H/+, or p53H/H mice were sacrificed at 2, 5, 7, and 15 h after 5 Gy of IR. Control mice were not irradiated. Mdm2 and p21 expression levels were measured in spleen RNA samples by real-time RT–PCR. Each bar depicts fold induction from triplicate samples after normalization to Gapdh. Wild-type samples without IR were set at 1.
To examine the stability of p53 in vivo with time, we irradiated four wild-type, four p53H/+, and four p53H/H mice with 5 Gy and sacrificed the mice at 2, 5, 7, and 15 h after IR (Fig. 3C,D). Nonirradiated mice were used as controls. Protein extracts were prepared from thymuses and spleens and p53 levels were assayed by Western blot analysis (Fig. 3C,D; data not shown). In wild-type spleens, p53 levels rose as expected at the 2-h time point (Fig. 3C,D). By 7 h post-irradiation, the wild-type p53 levels had returned to basal levels. p53H/H mice also showed increased mutant p53 levels at the 2-h time point, but in contrast to wild-type p53, p53R172H levels reached a plateau at the 5-h time point. At 15 h, p53R172H levels were still elevated. A study of p53H/+ mice revealed that p53 levels were also elevated at the 2-h time point, similar to wild-type and mutant p53 in response to radiation (Fig. 3C,D). However, the levels of a mixed wild-type/mutant p53 tetramer remained higher for a longer time than wild-type p53, but not as long as mutant p53, confirming the mixed nature of this p53 tetramer. These data indicate that p53R172H is stabilized like wild-type p53 in response to DNA damage. However, unlike wild-type p53, p53R172H remains stable for a significantly longer time.
One possible explanation for the increased stability of mutant p53 is that Mdm2 levels do not increase in the presence of mutant p53 and that basal Mdm2 levels cannot compensate for the increased p53. We therefore analyzed RNA samples of spleens from control and irradiated wild-type, p53H/+, and p53H/H mice for Mdm2 expression at 0, 2, 5, 7, and 15 h after 5 Gy of IR. Mdm2 mRNA levels increased with IR treatment and were highest in wild-type mice (Fig. 3E). p53H/+ mice also showed increased Mdm2 mRNA levels in response to IR but were about half that of wild-type mice. p53H/H mice showed basal levels of Mdm2 expression with no change after IR. Another p53 target, p21, followed a similar pattern of expression (Fig. 3E). Thus, wild-type mice activated Mdm2 after IR, but mutant p53 mice were compromised for Mdm2 expression.
Both p16INK4a/Rb and p53 are critical tumor suppressor pathways that engage in cross-talk at different levels (Sherr and McCormick 2002). Loss of p16INK4a results in increased Cyclin-D1/cdk activity, increased phosphorylation of Rb, and release of the E2F1 transcription factor, ultimately resulting in stabilization and increased activity of wild-type p53 in normal cells. To directly address the role of p16INK4a in mutant p53 stability, we crossed p53H/H mice to p16INK4a−/− mice that retain the p19ARF allele (Sharpless et al. 2001) to generate p53H/H p16INK4a−/− mice. Some cells from normal tissues from these mice—including bone marrow, intestine, and spleen—showed detectable levels of mutant p53 by IHC (Fig. 1B; Supplemental Fig. S1). These data indicate that p16INK4a also regulates mutant p53 levels in vivo. The cohort of p53H/H p16INK4a−/− mice developed tumors with an earlier age of onset (median survival 114 d; P < 0.0001) than p53H/H mice, demonstrating cooperation between this p53 mutation and loss of p16INK4a in tumorigenesis (Fig. 2C). Of 22 tumors that have developed thus far in p53H/H p16INK4a−/− mice, 13 were lymphomas, eight were sarcomas, and one was identified as a seminoma. Three of the eight (33%) sarcomas (one hemangiosarcoma and two histiocytic sarcomas) metastasized, whereas metastasis was never observed in p53H/H or p16INK4a−/− mice.
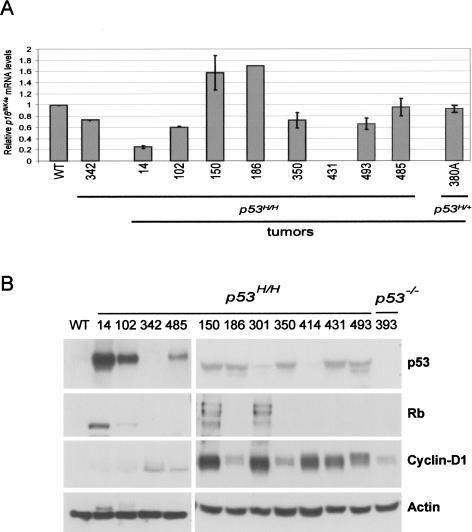
Since the absence of p16INK4a stabilized mutant p53 in normal tissues, we asked whether p16INK4a loss contributed to mutant p53 stability in tumors from p53H/H mice. All 10 tumors from p53H/H p16INK4a−/− mice examined were positive for mutant p53 by IHC (data not shown). We next probed the expression of p16INK4a in tumors from p53H/H mice with stable p53 by quantitative RT–PCR to determine if p16INK4a loss was a common event in the genesis of these tumors. Five of eight lymphomas from p53H/H mice showed decreased p16INK4a mRNA levels when compared with wild-type thymus control (Fig. 4A). One of these tumors (#431) completely lacked p16INK4a expression. In summary, some tumors with stable mutant p53 were compromised for p16INK4a expression.
Figure 4.
p16INK4a, Cyclin-D1, and Rb status in lymphomas from homozygous p53H/H mutant mice. (A) Quantitative RT–PCR analysis was performed to examine expression of p16INK4a in tumors from p53H/H mice. Each data point is depicted as the fold induction from triplicate samples after normalization to Gapdh. Tumor sample from mouse #380A did not have detectable p53 levels and is used as a negative control. Sample #342 is from a histologically normal thymus from a p53H/H mouse. (B) Western blot analysis of tumors from p53H/H mice using p53, Rb, and Cyclin-D1 antibodies.
Lastly, loss of p16INK4a is not the only defect in the Rb pathway seen in human tumors. Increased Cyclin-D1/cdk levels and phosphorylation of Rb are also observed and lead to release of the E2F1 transcription factor, resulting in stabilization and increased activity of wild-type p53. We therefore examined tumor samples from p53H/H mice to determine if they had alterations in Cyclin-D1 levels or Rb phosphorylation. Tumors with detectable p53R172H showed up-regulation of Cyclin-D1 (eight of 10 tumors), and phosphorylation of Rb (two of 10 tumors) (Fig. 4B). As a control, a thymus from a wild-type mouse had undetectable levels of both proteins on Western blots. We also examined a thymus from a p53H/H mouse (#342) prior to the onset of cancer and a p53-null tumor. These showed slightly elevated levels of Cyclin-D1. In summary, most tumors with stable mutant p53 had a defect in the Rb pathway.
Discussion
Our studies indicate that p53H/H mice do not have detectable mutant p53 in normal cells. The data also show that Mdm2 and DNA damage regulate mutant p53 as well as wild-type p53. DNA damage signals, however, resulted in a longer half-life for mutant p53. Since mutant p53 cannot activate Mdm2, the basal levels of Mdm2 may be insufficient to reduce mutant p53 levels as quickly as wild-type p53. Additionally, perhaps a circuit exists in cells that measures the wild-type p53 response. In the absence of wild-type p53 activity, signals inducing p53 persist but stabilize mutant p53. Another possibility is that once stabilized, mutant but not wild-type p53 interacts with other proteins that modify its stability. Mutant but not wild-type p53, for example, binds p53 family members p63 and p73 (Lang et al. 2004; Lozano 2007). These studies imply that secondary alterations are required to stabilize mutant p53 in tumors and are supported by the observation that 11% of human breast cancers have p53 missense mutations that are not IHC-positive (Sjogren et al. 1996).
The second observation from these studies is that the metastatic gain-of-function phenotype invariably associates with a stabilized mutant p53. The survival curves of mice inheriting two different mutant p53 alleles versus a p53-null allele do not vary, presumably because of rate-limiting events required to stabilize that mutant p53 (Lang et al. 2004; Olive et al. 2004). In contrast, when mutant p53 is stabilized by loss of Mdm2 or p16INK4a, these mice die earlier than p53 mutant mice and develop a metastatic phenotype. These data categorize tumors into two classes: less aggressive tumors with an unstable mutant p53 comparable with tumors lacking p53, and more aggressive tumors that accumulate alterations that stabilize mutant p53. The earlier tumor-specific alterations occur to stabilize mutant p53, the greater the opportunity for metastasis.
One other pathway that impacts p53 is the p16INK4a/Rb pathway. In a variety of human tumors, both tumor suppressor genes p16INK4a and p53 are commonly inactivated by different mechanisms (Levine 1997; Rocco and Sidransky 2001). p16INK4a loss leads to stable wild-type p53; conversely, overexpression of p16INK4a decreases p53 stability through Rb signaling in human mammary epithelial cells (Zhang et al. 2006). In our studies, mutant p53 mice lacking p16INK4a had stable mutant p53 in some but not all cells. The same is true in tissues analyzed with Mdm2 loss, suggesting cell context also contributes to mutant p53 stability. These observations indicate that while the same signals regulate wild-type and mutant p53, different cell types stabilize p53 by different mechanisms.
In conclusion, our data show that mutant p53 is regulated by the same signals as wild-type p53, and suggest that other pathways or signals may feed into this pathway to affect the stability of mutant and wild-type p53. The importance of this study cannot be overemphasized. Drugs that disrupt the interaction of Mdm2 with wild-type p53 will also disrupt interaction of Mdm2 with mutant p53 and thereby have adverse effects on tumor metastases in vivo. Moreover, chemotherapeutic agents that stabilize wild-type p53 may also stabilize mutant p53. Our studies raise the possibility that eliminating mutant p53 stability (a more feasible option than making mutant p53 wild type) may suppress the metastatic phenotype.
Material and methods
Mice
Mdm2 heterozygous mice were crossed to p53H/H (Lang et al. 2004) to generate p53H/+ Mdm2+/−, which we intercrossed to generate p53H/H Mdm2−/− and control littermates. These mice were maintained on a mixed 129S6/SvEv × C57BL/6 background. To determine mouse genotypes, PCR analysis was performed on tail DNA using published primer sets for Mdm2 (Montes de Oca Luna et al. 1995), p53H/H (Lang et al. 2004), and p53−/− (Jacks et al. 1994). p16INK4a−/− mice (Sharpless et al. 2001) were crossed to p53H/H mice to generate p53H/+ p16INK4a+/− mice, which were intercrossed to generate p53H/H p16INK4a−/− mice. Mice of different genotypes were treated with 5 Gy radiation (Cs137 source). All animal studies were approved by the Institutional Animal Care and Use Committee.
Statistics
Log-rank tests and Kaplan-Meier analyses were performed to assay statistical differences. A factor was considered statistically significant if it had a two-sided P-value of <0.05.
p53 immunohistochemical assay
Immunohistochemical analysis of p53 was performed as described previously (Evans et al. 2001) on 5-μm paraffin-embedded sections using a commercially available polyclonal rabbit antibody (CM5; Vector Laboratories, VP-P956) at 1:200 dilution overnight at 4°C.
Western blotting
Tissue lysates (80 μg of total protein) were prepared from either the thymus or spleen. Proteins were resolved by SDS-PAGE and transferred for 1 h and 30 min to PVDF membrane (Bio-Rad, 162-0177) using a semidry transfer apparatus. After blocking for 1 h at room temperature in 7.5% skim milk in 1× PBS–0.1% Tween (PBS-T), membranes were incubated with p53 (CM5; Vector Laboratories, VP-P956; 1:500 dilution), Cyclin-D1 (72-13G; Santa Cruz Biotechnologies, SC-450; 1:1000 dilution), and Rb (Ab-1; Oncogene, OP-28; 1:1000) primary antibodies overnight at 4°C. The next day, the membranes were washed with PBS-T, incubated with horseradish peroxidase-conjugated secondary antibodies, and visualized with ECL plus (GE Healthcare, RPN2132). As a loading control, we used anti-β-actin (Sigma, A5441; 1:2000) or anti-vinculin (Sigma, V4505; 1:1000) antibodies.
RT–PCR
RNA was isolated from tumor or spleen samples using the RNeasy kit (Qiagen, 74104). First-strand DNA was generated by using First-strand cDNA synthesis kit from GE Healthcare (27-9261-01). For PCR, the primer sequences to amplify p21, p16INK4a, and Gapdh were described previously (Burns et al. 2001; Gannon et al. 2002; Iwakuma et al. 2004). Primer sequences for Mdm2 were forward 5′-AGCGGGAGACCGTCC GGACAC-3′ and reverse 5′-AGAGTCTCTTGTTCCGAAGC 3′. mRNA expression was normalized to the expression of Gapdh in each reaction.
Acknowledgments
We thank Sandy Chang and Ron A. DePinho for providing the p16INK4a mice. We also thank Angelito De Villa and Maurice J. Dufilho IV for histology, and Arlette A. Audiffred, Ana C. Elizondo-Fraire, and Peirong Yang for technical support. This study was supported by a NIH grants CA34936 and U01 CA04002 to G.L. S.M.P. is supported by a Ruth L. Kirschstein NRSA fellowship (F32CA119616) and is a recipient of the Dowdy P. Hawn post-doctoral award. G.L. is the recipient of the Mattie Allen Fair Research Chair.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1662908.
References
- Appella E., Anderson C.W. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 2001;268:2764–2772. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- Brooks C.L., Gu W. Ubiquitination, phosphorylation and acetylation: The molecular basis for p53 regulation. Curr. Opin. Cell Biol. 2003;15:164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- Burns T.F., Bernhard E.J., El-Deiry W.S. Tissue specific expression of p53 target genes suggests a key role for KILLER/DR5 in p53-dependent apoptosis in vivo. Oncogene. 2001;20:4601–4612. doi: 10.1038/sj.onc.1204484. [DOI] [PubMed] [Google Scholar]
- Canman C.E., Lim D.S., Cimprich K.A., Taya Y., Tamai K., Sakaguchi K., Appella E., Kastan M.B., Siliciano J.D. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- Chavez-Reyes A., Parant J.M., Amelse L.L., de Oca Luna R.M., Korsmeyer S.J., Lozano G. Switching mechanisms of cell death in mdm2- and mdm4-null mice by deletion of p53 downstream targets. Cancer Res. 2003;63:8664–8669. [PubMed] [Google Scholar]
- Chuikov S., Kurash J.K., Wilson J.R., Xiao B., Justin N., Ivanov G.S., McKinney K., Tempst P., Prives C., Gamblin S.J., et al. Regulation of p53 activity through lysine methylation. Nature. 2004;432:353–360. doi: 10.1038/nature03117. [DOI] [PubMed] [Google Scholar]
- Dittmer D., Pati S., Zambetti G., Chu S., Teresky A.K., Moore M., Finlay C., Levine A.J. Gain of function mutations in p53. Nat. Genet. 1993;4:42–46. doi: 10.1038/ng0593-42. [DOI] [PubMed] [Google Scholar]
- Eischen C.M., Weber J.D., Roussel M.F., Sherr C.J., Cleveland J.L. Disruption of the ARF–Mdm2–p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes & Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans S.C., Viswanathan M., Grier J.D., Narayana M., El-Naggar A.K., Lozano G. An alternatively spliced HDM2 product increases p53 activity by inhibiting HDM2. Oncogene. 2001;20:4041–4049. doi: 10.1038/sj.onc.1204533. [DOI] [PubMed] [Google Scholar]
- Ferbeyre G., de Stanchina E., Lin A.W., Querido E., McCurrach M.E., Hannon G.J., Lowe S.W. Oncogenic ras and p53 cooperate to induce cellular senescence. Mol. Cell. Biol. 2002;22:3497–3508. doi: 10.1128/MCB.22.10.3497-3508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores E.R., Sengupta S., Miller J.B., Newman J.J., Bronson R., Crowley D., Yang A., McKeon F., Jacks T. Tumor predisposition in mice mutant for p63 and p73: Evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7:363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- Gannon G., Mandriota S.J., Cui L., Baetens D., Pepper M.S., Christofori G. Overexpression of vascular endothelial growth factor-A165 enhances tumor angiogenesis but not metastasis during β-cell carcinogenesis. Cancer Res. 2002;62:603–608. [PubMed] [Google Scholar]
- Giaccia A.J., Kastan M.B. The complexity of p53 modulation: Emerging patterns from divergent signals. Genes & Dev. 1998;12:2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- Gu W., Roeder R.G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Hall P.A., Lane D.P. p53 in tumour pathology: Can we trust immunohistochemistry?—Revisited! J. Pathol. 1994;172:1–4. doi: 10.1002/path.1711720103. [DOI] [PubMed] [Google Scholar]
- Haupt Y., Maya R., Kazaz A., Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Helton E.S., Chen X. p53 modulation of the DNA damage response. J. Cell. Biochem. 2007;100:883–896. doi: 10.1002/jcb.21091. [DOI] [PubMed] [Google Scholar]
- Hermeking H., Eick D. Mediation of c-Myc-induced apoptosis by p53. Science. 1994;265:2091–2093. doi: 10.1126/science.8091232. [DOI] [PubMed] [Google Scholar]
- Honda R., Tanaka H., Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- Horn H.F., Vousden K.H. Coping with stress: Multiple ways to activate p53. Oncogene. 2007;26:1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- Iwakuma T., Parant J.M., Fasulo M., Zwart E., Jacks T., de Vries A., Lozano G. Mutation at p53 serine 389 does not rescue the embryonic lethality in mdm2 or mdm4 null mice. Oncogene. 2004;23:7644–7650. doi: 10.1038/sj.onc.1207793. [DOI] [PubMed] [Google Scholar]
- Jacks T., Remington L., Williams B.O., Schmitt E.M., Halachmi S., Bronson R.T., Weinberg R.A. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Jones S.N., Roe A.E., Donehower L.A., Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- Kamb A., Gruis N.A., Weaver-Feldhaus J., Liu Q., Harshman K., Tavtigian S.V., Stockert E., Day R.S., Johnson B.E., Skolnick M.H. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- Kamijo T., Weber J.D., Zambetti G., Zindy F., Roussel M.F., Sherr C.J. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl. Acad. Sci. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan M.B., Onyekwere O., Sidransky D., Vogelstein B., Craig R.W. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- Kubbutat M.H., Jones S.N., Vousden K.H. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Lang G.A., Iwakuma T., Suh Y.A., Liu G., Rao V.A., Parant J.M., Valentin-Vega Y.A., Terzian T., Caldwell L.C., Strong L.C., et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Lavin M.F., Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006;13:941–950. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- Levine A.J. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Lozano G. The oncogenic roles of p53 mutants in mouse models. Curr. Opin. Genet. Dev. 2007;17:66–70. doi: 10.1016/j.gde.2006.12.003. [DOI] [PubMed] [Google Scholar]
- McDonnell T.J., de Montes Oca Luna R., Cho S., Amelse L.L., Chavez-Reyes A., Lozano G. Loss of one but not two mdm2 null alleles alters the tumour spectrum in p53 null mice. J. Pathol. 1999;188:322–328. doi: 10.1002/(SICI)1096-9896(199907)188:3<322::AID-PATH372>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Medema R.H., Herrera R.E., Lam F., Weinberg R.A. Growth suppression by p16ink4 requires functional retinoblastoma protein. Proc. Natl. Acad. Sci. 1995;92:6289–6293. doi: 10.1073/pnas.92.14.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek D.W. Post-translational modification of p53. Semin. Cancer Biol. 1994;5:203–210. [PubMed] [Google Scholar]
- de Montes Oca Luna R., Wagner D.S., Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- Nobori T., Miura K., Wu D.J., Lois A., Takabayashi K., Carson D.A. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature. 1994;368:753–756. doi: 10.1038/368753a0. [DOI] [PubMed] [Google Scholar]
- Olive K.P., Tuveson D.A., Ruhe Z.C., Yin B., Willis N.A., Bronson R.T., Crowley D., Jacks T. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Palmero I., Pantoja C., Serrano M. p19ARF links the tumour suppressor p53 to Ras. Nature. 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- Pomerantz J., Schreiber-Agus N., Liegeois N.J., Silverman A., Alland L., Chin L., Potes J., Chen K., Orlow I., Lee H.W., et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- Prives C., Hall P.A. The p53 pathway. J. Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Quelle D.E., Zindy F., Ashmun R.A., Sherr C.J. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- Rocco J.W., Sidransky D. p16(MTS-1/CDKN2/INK4a) in cancer progression. Exp. Cell Res. 2001;264:42–55. doi: 10.1006/excr.2000.5149. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K., Herrera J.E., Saito S., Miki T., Bustin M., Vassilev A., Anderson C.W., Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes & Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M., Lin A.W., McCurrach M.E., Beach D., Lowe S.W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Sharpless N.E., Bardeesy N., Lee K.H., Carrasco D., Castrillon D.H., Aguirre A.J., Wu E.A., Horner J.W., DePinho R.A. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- Sherr C.J. Tumor surveillance via the ARF–p53 pathway. Genes & Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- Sherr C.J., McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- Shieh S.Y., Ikeda M., Taya Y., Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- Sigal A., Rotter V. Oncogenic mutations of the p53 tumor suppressor: The demons of the guardian of the genome. Cancer Res. 2000;60:6788–6793. [PubMed] [Google Scholar]
- Sjogren S., Inganas M., Norberg T., Lindgren A., Nordgren H., Holmberg L., Bergh J. The p53 gene in breast cancer: Prognostic value of complementary DNA sequencing versus immunohistochemistry. J. Natl. Cancer Inst. 1996;88:173–182. doi: 10.1093/jnci/88.3-4.173. [DOI] [PubMed] [Google Scholar]
- Song H., Hollstein M., Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat. Cell Biol. 2007;9:573–580. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- Takagi M., Absalon M.J., McLure K.G., Kastan M.B. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Vousden K.H. Outcomes of p53 activation—Spoilt for choice. J. Cell Sci. 2006;119:5015–5020. doi: 10.1242/jcs.03293. [DOI] [PubMed] [Google Scholar]
- Wagner A.J., Kokontis J.M., Hay N. Myc-mediated apoptosis requires wild-type p53 in a manner independent of cell cycle arrest and the ability of p53 to induce p21waf1/cip1. Genes & Dev. 1994;8:2817–2830. doi: 10.1101/gad.8.23.2817. [DOI] [PubMed] [Google Scholar]
- Weisz L., Oren M., Rotter V. Transcription regulation by mutant p53. Oncogene. 2007;26:2202–2211. doi: 10.1038/sj.onc.1210294. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Xiong Y., Yarbrough W.G. ARF promotes MDM2 degradation and stabilizes p53: ARF–INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- Zhang J., Pickering C.R., Holst C.R., Gauthier M.L., Tlsty T.D. p16INK4a modulates p53 in primary human mammary epithelial cells. Cancer Res. 2006;66:10325–10331. doi: 10.1158/0008-5472.CAN-06-1594. [DOI] [PubMed] [Google Scholar]