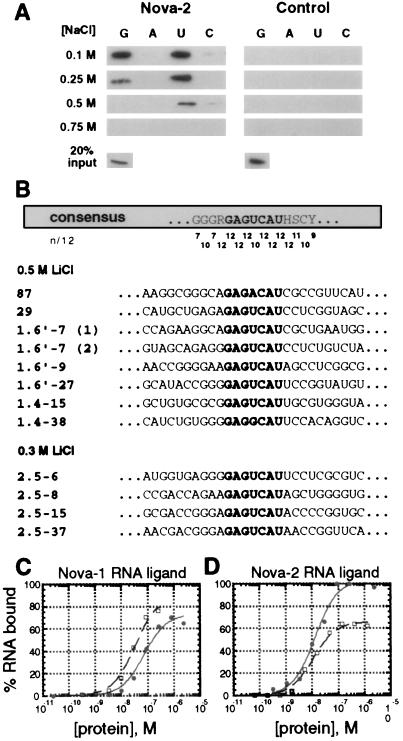
Figure 4.
Nova-2 is an RNA-binding protein. (A) Analysis of Nova-2 RNA-binding ability to ribohomopolymer RNA in varying salt concentrations. N2FP (5 μg) or an irrelevant T7 epitope-tagged and histidine-tagged control protein was incubated with poly(G), poly(A), poly(U), and poly(C)-Sepharose ribohomopolymer beads in either 0.1 M, 0.25 M, 0.5 M, or 0.75 M NaCl. After being washed, protein bound to the ribohomopolymers was analyzed by Western blot analysis with an anti-T7 antibody (Novagen). Twenty percent of the input protein is shown (Lower). (B) Affinity elution-based RNA selection identifies a Nova-2 consensus binding sequence. Alignment of 12 Nova-2 RNA ligands selected in two independent Nova-2 selection experiments (in 0.5 and 0.3 M LiCl) is shown. A consensus sequence is shown at the top, with the GAGUCAU motif highlighted. The match to the consensus, scored as the number of exact matches of 12 sequences (n/12), is shown for each position. Bases in RNA ligands with identity to the GAGUCAU motif are also shown highlighted. (C and D). Direct comparisons of Nova-1 and Nova-2 binding to selected RNA ligands. Nitrocellulose filter binding assays were performed to examine Nova-1 (open squares) and Nova-2 (solid circles) binding to RNA ligands mSB2 (C), a 21-nt RNA containing the Nova-1 consensus binding motif [UCAU(N)0–2]3 (19); and 87 (D), a 96-nt RNA ligand isolated by Nova-2 RNA selection (Fig. 4B). RNA ligand mSB2 bound to Nova-1 and Nova-2 with calculated Kd values of 33 ± 5 nM and 73 ± 13 nM, respectively (calculated by least squares fit; ref. 33). Similar results were found with the 96 nt SB2 Nova-1 RNA ligand (data not shown). RNA ligand 87 bound to Nova-1 and Nova-2 with calculated Kd values of 13 ± 2.5 nM and 11 ± 1.5 nM, respectively. Maximal Nova-1 binding to RNA ligand 87 was observed at ≈65% of the total RNA, while maximal Nova-2 binding was 100%. Similar results were found with the Nova-2 RNA ligand 29 (data not shown). Neither Nova protein demonstrated significant binding to a random RNA ligand isolated from the original RNA selection pool, and an irrelevant T7 epitope-tagged recombinant control protein did not demonstrate significant binding to any of the RNA ligands (calculated Kd values > 1 μM, data not shown).