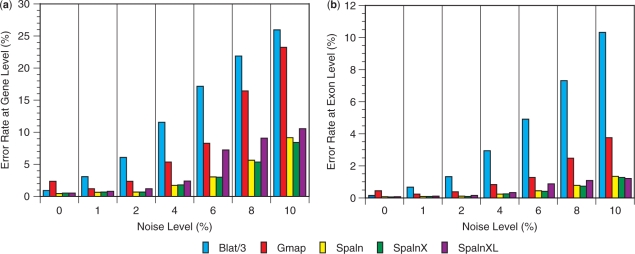
Figure 4.
Accuracy in mapping and alignment of CDS sequences onto the human or mouse genomic sequence. Error rates at the gene level (a) and at the exon level (b) are shown as functions of artificially introduced noise levels. The actual error rates of Blat are three times as large as those indicated by the bar heights. SpalnXL indicates that ‘-yX –LS’ options were applied to Spaln at run time. Each error rate is the mean of six trials, the numerical value and the SD of which are shown in Table S9 with other information.