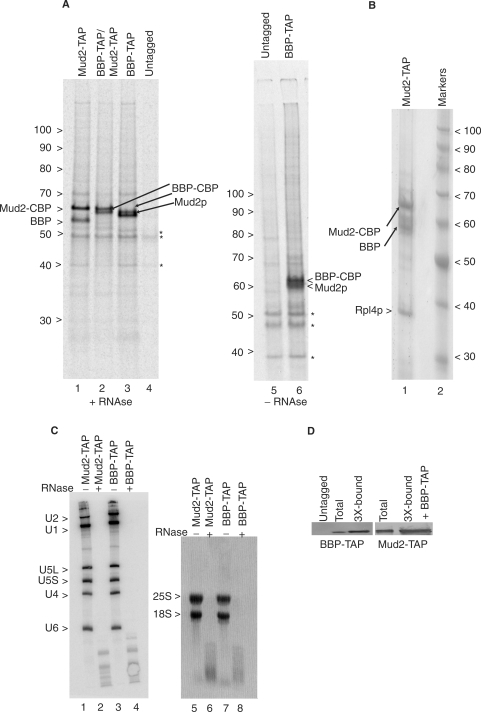
Figure 1.
Tandem affinity purification of BBP–TAP and Mud2–TAP. (A) Autoradiogram of a 7% polyacrylamide gel of metabolically labeled proteins selected by sequential IgG agarose and calmodulin agarose chromatography from yeast that express the indicated TAP-tagged gene constructs as genomic integrants. The untagged lane shows background proteins (asterisks). Samples in lanes 1–4 were pre-treated with RNase A prior to TAP selection while samples 5–6 were not. The numbers at the left indicate the positions of unlabeled protein molecular weight markers. Bands corresponding to the untagged Mud2p and BBP proteins and the proteins with the residual CBP tags are indicated. (B) Coomassie blue stain of unlabeled Mud2-TAP and co-purifying proteins. Lane 2 shows the migration of protein molecular weight markers. (C) Yeast used in panel a treated (+) or not (−) with RNase A hybridized with probes specific for the spliceosomal snRNAs (lanes 1–4) or stained with ethidium bromide for the 25S and 18S ribosomal RNAs (lanes 5–8). (D) Single-step recovery of BBP–TAP and Mud2–TAP by TAP by calmodulin agarose chromatography. To avoid band distortion due to sample overloading, the unfractionated (total) protein lanes contain 1/3 the equivalent amount of sample. Untagged = calmodulin agarose recovered material recovered from an untagged extract.