Abstract
Transcription factors regulate tissue patterning and cell fate determination during development; however, expression of early regulators frequently abates upon differentiation, suggesting that they may also play a role in maintaining an undifferentiated phenotype. The transcription factor paired box 3 (Pax3) is expressed by multipotent neural crest precursors and is implicated in neural crest disorders in humans such as Waardenburg syndrome. Pax3 is required for development of multiple neural crest lineages and for activation of lineage-specific programs, yet expression is generally extinguished once neural crest cells migrate from the dorsal neural tube and differentiate. Using a murine Cre-inducible system, we asked whether persistent Pax3 expression in neural crest derivatives would affect development or patterning. We found that persistent expression of Pax3 in cranial neural crest cells resulted in cleft palate, ocular defects, malformation of the sphenoid bone, and perinatal lethality. Furthermore, we demonstrated that Pax3 directly regulates expression of Sostdc1, a soluble inhibitor of bone morphogenetic protein (BMP) signaling. Persistent Pax3 expression renders the cranial crest resistant to BMP-induced osteogenesis. Thus, one mechanism by which Pax3 maintains the undifferentiated state of neural crest mesenchyme may be to block responsiveness to differentiation signals from the environment. These studies provide in vivo evidence for the importance of Pax3 downregulation during differentiation of multipotent neural crest precursors and cranial development.
Introduction
Among the most intriguing discoveries in developmental biology in the last century has been the identification of powerful developmentally regulated transcription factors that regulate patterning and cell-fate determination during embryogenesis. Expression of myogenic basic helix-loop-helix transcription factors, for example, is required for skeletal muscle differentiation and can induce myogenic differentiation in naive cells (1–4). Homeobox genes specify anterior-posterior identity and are frequently required for patterning and differentiation of organs and tissues throughout the body (5). Gene inactivation studies in mice and other organisms have verified the critical role played by innumerable transcriptional regulators during embryogenesis. Many of these factors are silenced or inactivated during embryonic development as differentiation proceeds, yet few studies have addressed the importance of this aspect of gene regulation. Is inactivation of specific developmental transcription factors required for differentiation and patterning to occur normally? Is turning off a gene just as important as turning it on? We sought to address these questions using an in vivo approach in mice.
Paired box 3 (Pax3) is a developmentally regulated transcription factor that is expressed in neural crest and somitic mesoderm (6) and is implicated in neural crest disorders, including the Splotch phenotype in mice and Waardenburg syndrome in humans. Pax3 is required for neural crest development and differentiation, since mutations in Pax3 result in a wide variety of neural crest defects, including small or absent dorsal root ganglia, melanocyte defects, and congenital heart disease related to deficient septation and smooth muscle development in the outflow tract of the heart (7, 8). In the skin, Pax3 expression abates during melanocyte differentiation of neural crest derivatives, but expression persists in a small population of undifferentiated melanocyte precursors, suggesting a role in stem cell biology (9). Pax3 is also required for some aspects of skeletal muscle development, where it is thought to act upstream of MyoD (10), and Pax3 is expressed by undifferentiated satellite cells, which can function as skeletal muscle stem cells (11, 12). Expression of a Pax3-forkhead (FKHR) fusion protein results in tumors of skeletal muscle in humans (rhabdomyosarcoma), and Pax3 itself is overexpressed in many cases of rhabdomyosarcoma that are not associated with the Pax3-FKHR translocation (13, 14).
Pax3 is a member of the paired-box family of transcription factors, which includes 9 members in mice and humans (7). Each member of the family is expressed in a tissue-specific pattern during development, and as in the case of Pax3, expression generally abates upon differentiation. In the eye, Pax6 plays a vital role since mutations in flies, mice, and humans result in severe ocular malformations or complete absence of the eye (15). Pax6 is required in retinal progenitor cells to maintain multipotency (16), and the persistence of Pax6-expressing cells in the adult eye in some lower organisms is associated with the ability to regenerate the eye upon injury (17, 18). In the hematopoietic system, Pax5 is expressed by pre–B cells and expression abates during further differentiation, while persistent expression is associated with some leukemias (19).
We have previously shown that Pax3 plays a cell-autonomous role in neural crest development, since transgenic rescue of Pax3 expression in neural crest is sufficient to rescue development and survival in a Pax3-null mouse (20). Direct transcriptional targets of Pax3 in neural crest are generally unknown, though it may regulate the cell-surface tyrosine kinase receptor c-ret in enteric ganglia precursors (21) and Mitf in melanoblasts (22, 23). Microarray analysis and chromatin immunoprecipitation studies have identified a number of other potential targets including TGF-β2 (24). A wide variety of neural crest defects have been associated with Pax3 deficiency, though some aspects of neural crest development are thought to be preserved in Pax3 mutants because of redundant functions of the closely related Pax7 gene, which is frequently coexpressed (25). Hence, Pax3/Pax7 double mutants have a more severe phenotype than either mutant alone. During development in the mouse, Pax3 expression initiates in the dorsal neural tube at about E8.0 followed shortly by expression of Pax7. By E10.5–E11, Pax3 expression is extinguished in neural crest cells as they migrate away from the neural tube, though expression by neuroblasts within the dorsal neural tube more gradually diminishes through gestation. In cranial regions, Pax3-expressing neural crest cells compose the mesenchymal cells of the maxillary and mandibular regions forming bone and muscle and contribute to the developing eyelids and periocular tissues.
Here, we describe studies that test the requirement for Pax3 to be inactivated during embryonic development. Using gene targeting and Cre-lox approaches, we have engineered mice in which Pax3 is activated normally during development, but expression persists beyond the time points at which it is normally shut off. This results in perinatal lethality, cleft palate, and additional defects. Further, using a combination of microarray and bioinformatics applications, we have identified what we believe is a novel downstream target of Pax3 regulation, the secreted bone morphogenetic protein (BMP) inhibitor sclerostin domain–containing 1 (Sostdc1). Persistent Pax3-mediated upregulation of Sostdc1 results in delayed osteogenesis, accounting for the lethal cleft palate phenotype. Thus, our results demonstrate the importance of gene inactivation during development.
Results
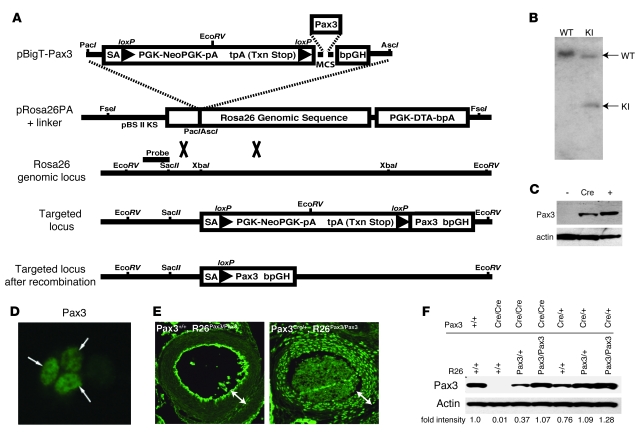
Targeting a Cre-inducible Pax3 cDNA to the Rosa locus.
The Rosa (Gt(ROSA)26Sor) locus had been successfully utilized to mediate widespread expression in the mouse embryo (26). We utilized a strategy designed to introduce the Pax3 cDNA into the Rosa locus immediately downstream of a loxP-flanked PGK-neo cassette that included stop codons in all 3 reading frames (Figure 1A). Thus, Pax3 would not be expressed unless Cre-mediated recombination removed the PGK-neo cassette, bringing Pax3 under the control of the Rosa locus regulatory elements. ES cells were targeted, and appropriate integration was confirmed by Southern blot (Figure 1B). Targeted (129X1/SvJ × 129S1/Sv) F1-derived R1 ES cells were capable of expressing Pax3 after infection with adenovirus encoding Cre recombinase (Figure 1, C and D), confirming appropriate targeting and Cre-mediated regulation. Correctly targeted ES cells were microinjected in C57BL/6 blastocysts, and chimeric progeny were established. After breeding to C57BL/6 mice, resulting heterozygotes were named R26Pax3/+ mice and subsequently derived homozygotes were named R26Pax3/Pax3 mice. Both heterozygotes and homozygotes appeared normal and were able to breed and thrive.
Figure 1. Pax3 knockin at Rosa26 locus.
(A) The murine Pax3 cDNA was inserted downstream of a PGK-neo cassette with a transcriptional stop signal flanked by 2 loxP sites in the pBigT plasmid containing the Rosa26 genomic sequence. Homologous recombination between the Rosa26Pax3 construct and the appropriate genomic locus resulted in a targeted allele where Pax3 is expressed only after Cre-mediated recombination. A probe for Southern blot confirmation of homologous recombination is indicated. (B) Southern blot analysis of WT and R26Pax3/+ EcoRV-digested genomic DNA from wild-type and targeted ES cells showing wild-type and knockin (KI) bands. (C) Western blot of lysates from uninfected (lane 1) mouse embryonic stem cells targeted with R26Pax3/+ or infected with Cre-adenovirus (lane 2) with Pax3 antibody. Pax3 protein from transfected 293T cells was used as a positive control (lane 3). (D) Cre-adenovirus–infected R26Pax3/+ ES cells immunostained with Pax3 antibody. Original magnification, ×100. (E) Immunohistochemistry to detect Pax3 protein in the smooth muscle layer of the aortic arch (arrows) of E16.5 embryos. Pax3 protein is detected in R26Pax3/Pax3 embryos only when Pax3Cre (right panel) is present. Endothelial and luminal staining is artifactual. Pax3 staining is nuclear. Original magnification, ×25. (F) Western blot to detect Pax3 protein in E11.5 whole-embryo lysates. Actin is shown as control, and relative pixel density is indicated below each lane.
Persistent expression of Pax3 in Pax3-expressing cells.
In order to activate Pax3 expression specifically in Pax3-expressing cells, we bred R26Pax3/+ mice with animals in which Cre recombinase had been targeted to the endogenous Pax3 locus (Pax3Cre/+ mice). Pax3Cre/+ mice have been previously described (27). In these animals, Cre recombinase expression faithfully mimics endogenous Pax3 expression, and Cre disrupts expression of Pax3 such that homozygous Pax3Cre/Cre animals phenocopy Splotch homozygous mutants. We obtained the expected number of compound heterozygous (Pax3Cre/+, R26Pax3/+) offspring, which appeared healthy. Male and female compound heterozygotes were subsequently inter-crossed, and the resulting embryos were examined at mid-gestation. 262 embryos between the ages of E11.5 and E17.5 were genotyped and roughly the expected number of animals of each genotype was observed (Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI33715DS1). However, no adult Pax3Cre/+, R26Pax3/Pax3 or Pax3Cre/Cre mice with or without R26Pax3 were observed past P2. Further, we found that although roughly the expected numbers of Pax3Cre/+, R26Pax3/Pax3 mice were born, none survived past P2, due to defects described below.
During embryogenesis, Pax3 expression initiates at approximately E8.0 in the dorsal neural tube and subsequently appears in presomitic mesoderm (6). Pax3 expression in noncranial neural crest normally abates shortly after delamination of neural crest cells from the neural tube, while expression persists slightly longer in cranial crest derivatives. By late gestation, the majority of Pax3 expression has disappeared, though expression in some Schwann cells (28) and satellite cells in muscle (12) persists into adulthood. Pax3 is required for subsets of neural crest and skeletal muscle development, and Pax3-deficient Splotch mutants display neural tube defects, deficiencies of neural crest, including congenital heart disease, and deficiencies of limb and other hypaxial muscles (7). By E13.5, neural crest–derived smooth muscle cells in the aortic arch did not express Pax3 protein (Figure 1E). However, in Pax3Cre/+, R26Pax3/+ embryos, Pax3 is easily detected in the smooth muscle layer of the aortic arch at E16.5 (not shown), and even higher levels of Pax3 expression are evident in Pax3Cre/+, R26Pax3/Pax3 embryos that carry 2 copies of the R26Pax3 allele (Figure 1E). Similar findings were observed in other Pax3-derived cell types, including dorsal neural tube, adrenal medulla, and skeletal muscle (data not shown). These results demonstrate that persistent Pax3 expression was effectively engineered in Pax3 derivatives.
At E11.5, we identified embryos of each of the expected genotypes (Supplemental Table 1), and we examined these for Pax3 expression by immunoblotting. As expected, we found little or no Pax3 protein expressed in Pax3Cre/Cre homozygotes, but the additional presence of 2 copies of the R26Pax3 allele restored Pax3 protein expression toward wild-type levels. Further, densitometric analysis demonstrated that regardless of the genetic locus of expression (Pax3 endogenous locus or Rosa26 locus), the presence of 1 copy of Pax3 resulted in an intermediate level of Pax3 protein, 2 copies resulted in near wild-type levels, and 3 copies (Pax3Cre/+, R26Pax3/Pax3) had only slightly elevated Pax3 protein levels (Figure 1F). Thus, persistent expression of Pax3 from the Rosa26 locus achieves levels comparable to those produced from the wild-type Pax3 locus at mid-gestation.
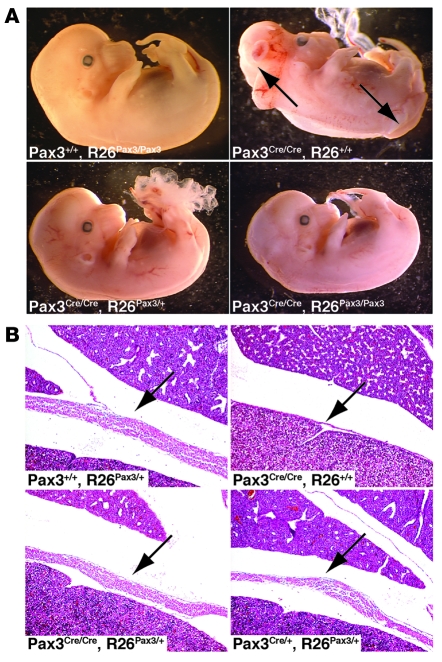
Rescue of developmental defects.
We have previously reported that Pax3Cre/Cre mice display neural tube and skeletal muscle defects similar to those seen in Pax3-deficient Splotch mice (27). We predicted that, if Pax3 protein expressed from the Cre-activated R26Pax3 allele was functional, this expression would be sufficient to rescue developmental defects in mice that lacked Pax3 derived from the endogenous locus. Pax3Cre/Cre, R26+/+ E13.5 embryos derived from intercrosses of Pax3Cre/+, R26Pax3/+ mice always displayed neural tube defects, either in the lumbar or cranial regions or both (Figure 2A). However, Pax3Cre/Cre embryos that also carried 1 or 2 alleles of R26Pax3 never displayed neural tube defects, indicating that Cre-dependent Pax3 expression from the Rosa locus was sufficient to rescue neural tube closure in embryos lacking Pax3 expression from the endogenous locus. Cre-mediated expression of Pax3 from the Rosa locus was also sufficient to rescue muscular development of the diaphragm (Figure 2B) and hind limb (Supplemental Figure 1), which is deficient in Pax3Cre/Cre embryos. However, we did not observe complete rescue of all hypaxial muscles in R26Pax3/Pax3, Pax3Cre/Cre embryos, since forelimb musculature remained hypoplastic (Supplemental Figure 1), perhaps because Cre-mediated activation of Pax3 expression from the Rosa locus was slightly delayed when compared with the onset of endogenous Pax3 expression in wild-type embryos. We conclude that expression from the Rosa locus in the manner described was able to rescue most, but not all, developmental functions of Pax3.
Figure 2. Persistent expression of Pax3 rescues Pax3-null defects.
(A) E13.5 littermates resulting from Pax3Cre/+, R26Pax3/+ intercrosses are shown. Note the open neural tube and exencephaly in the Pax3Cre/Cre, R26+/+ embryo (arrows, upper right panel) that are rescued in the embryos persistently expressing Pax3 (lower panels). (B) H&E-stained sagittal sections of E17.5 littermates show the diaphragm (arrows), which is thin and lacking muscle in Pax3Cre/Cre, R26+/+ embryos (upper right panel) but restored in the embryos persistently expressing Pax3 (lower panels). Original magnification, ×6.3.
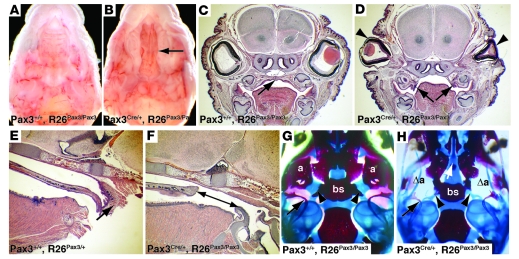
Cleft palate in mice persistently expressing Pax3.
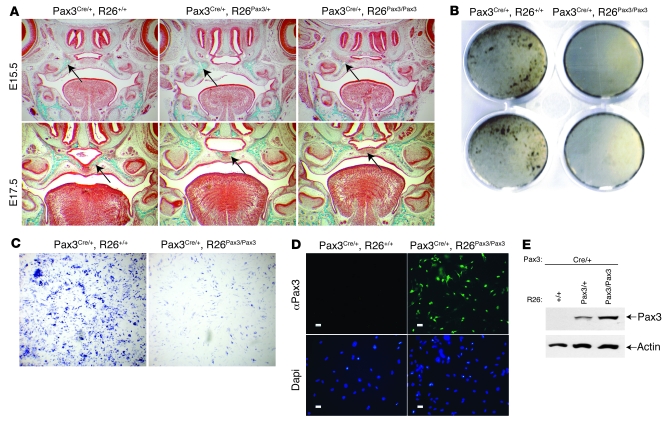
Although Pax3Cre/+, R26Pax3/+ mice survived, we never observed Pax3Cre/+, R26Pax3/Pax3 mice beyond P2 (Supplemental Table 1). This suggested dosage sensitivity to persistent expression of Pax3 in Pax3 derivatives. Pax3Cre/+, R26Pax3/Pax3 mice were found alive at birth, but succumbed shortly thereafter and failed to suckle successfully. These mice all displayed cleft palate. Obvious gross palatal defects were seen in 5 of the 16 newborn Pax3Cre/+, R26Pax3/Pax3 pups examined (Figure 3, A–D), and in the cases where gross defects were not apparent, analysis of sagittal sections revealed significant foreshortening of the palate (Figure 3, E and F). Therefore, all Pax3Cre/+, R26Pax3/Pax3 mice displayed significant palate defects. In addition, we also noted subtle palate defects upon histologic analysis in many mice heterozygous for the persistent Pax3 allele (Pax3Cre/+, R26Pax3/+) in which the palate was mildly shortened (data not shown).
Figure 3. Persistent Pax3 expression causes cleft palate.
(A and B) P0 heads are shown with the lower jaw removed to reveal the palate. Persistent Pax3 expression in the Pax3Cre/+, R26Pax3/Pax3 mouse (B) results in cleft palate (arrow) compared with a control Pax3+/+, R26Pax3/Pax3 littermate (A). (C and D) Frontal sections through E16.5 heads reveal properly raised and fused palatal shelves in control (arrow, C) embryos. Mutant (Pax3Cre/+, R26Pax3/Pax3) littermates display a cleft with palatal shelves that fail to lift (arrows, D) as well as an absence of eyelids (arrowheads, D). (E and F) Sagittal sections of P0 heads indicate that control palates extend and contact the epiglottis in the control (arrow, E) but are shortened in the mutant (arrow, F). (G and H) Alizarin red/Alcian blue–stained P0 heads with the mandibles removed reveal that mutant (Pax3Cre/+, R26Pax3/Pax3) pups have a complete absence of the secondary palate (white arrow, center, H) as well as missing alisphenoid (Δa) bones, deformed pterygoid processes of the basisphenoid bone (arrowheads), and thinned tympanic rings (black arrow) compared with control (G) littermates. a, alisphenoid; bs, basisphenoid. Original magnification, ×6 (A, B); ×12.5 (C, D); ×15.5 (E, F); ×7.5 (G, H).
Skeletal preparations confirmed severe palate defects in Pax3Cre/+, R26Pax3/Pax3 mice (Figure 3, G and H) and revealed additional abnormalities of the bony derivatives of cranial neural crest. Pax3Cre/+, R26Pax3/Pax3 mice displayed additional craniofacial malformations such as hypoplastic or absent alisphenoid bones and pterygoid processes of the basisphenoid bones as well as drastically thinned tympanic bones (Figure 3, G and H), all of which are at least partially derived from cells of neural crest origin (29–33). These results suggest that inactivation of Pax3 is critical for proper development of some cranial neural crest–derived osseous structures. Further, all Pax3Cre/+, R26Pax3/Pax3 mice (26 of 26) had defects in eyelid development of varying severity, from eyelids that failed to fuse to completely missing eyelids (Figure 3, C and D, and Supplemental Figure 2). Quail-chick chimera experiments have demonstrated that neural crest cells migrate to and intermingle with mesodermal cells to contribute to both upper and lower eyelid development (34).
We sought to confirm these results by performing an independent experiment in which persistent Pax3 expression was activated using a transgenic mouse with well-characterized expression of Cre recombinase that is limited to neural crest (29, 35). Using Wnt1-Cre–driven persistent expression of Pax3, we observed identical morphological defects involving the palate, eyelids, and cranial bones in Wnt1Cre, R26Pax3/Pax3 mice compared with Pax3Cre/+, R26Pax3/Pax3, and all 15 Wnt1Cre, R26Pax3/Pax3 mice examined were abnormal. We also observed gross cleft palate in 1 of 13 mice that carried only 1 copy of the persistent Pax3 allele (Wnt1Cre, R26Pax3/+) at P0. These results confirm that persistent expression of Pax3 specifically in neural crest–derived cells is responsible for the observed defects.
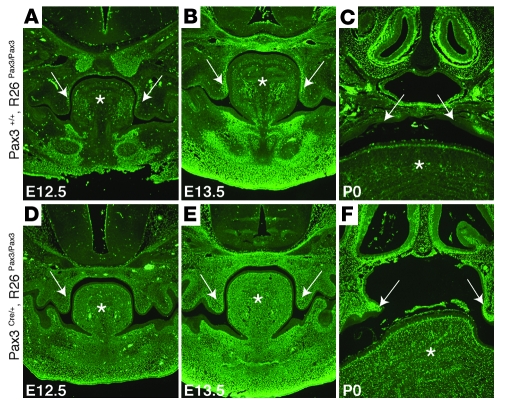
Immunohistological examination demonstrated that endogenous Pax3 protein is present in the developing palatal shelves at E12.5 and E13.5 during palatal shelf growth and expansion (Figure 4). At this stage, Pax3 is detected in a domain that is restricted to the inner surface of palate mesenchyme (Figure 4, A and B) but is no longer expressed in the fused palate at birth (Figure 4C). In contrast, Pax3 is robustly expressed in an expanded domain encompassing the entire palate mesenchyme as well as the entire tongue and developing mandible of Pax3Cre/+, R26Pax3/Pax3 mice at E12.5 and E13.5 (Figure 4, D and E), indicating that this tissue consists of cells that are neural crest derivatives, which had expressed endogenous Pax3 at one point. Persistent Pax3 expression is not seen in palate epithelium or surface ectoderm and is restricted to neural crest–derived tissues. Further, Pax3 protein is still detected in the palates of Pax3Cre/+, R26Pax3/Pax3 mice at birth (Figure 4F), when control mice no longer have detectable Pax3 in this region. These results indicate that the persistent Pax3-expressing neural crest cells migrate to the proper craniofacial locations where they survive to populate the mature structures. Analysis of the developing palate revealed no changes in proliferation (data not shown).
Figure 4. Endogenous and persistent Pax3 is expressed in the palate.
Frontal sections through the palatal region of E12.5 (A and D), E13.5 (B and E), and P0 (C and F) embryos are shown. Endogenous Pax3 is expressed in Pax3+/+, R26Pax3/Pax3 embryos in the palatal shelves at E12.5 (A) and E13.5 (B) along the inner surface (arrows) as well as in the tongue (*) and mandible. At birth, Pax3 expression in the palate has abated in control embryos (F). Pax3Cre/+, R26Pax3/Pax3 mice have much higher levels of Pax3 in the palate (arrows) and tongue (*) compared with control at both E12.5 (D) and E13.5 (E), with expression expanded to the entire palate mesenchyme, tongue, and developing mandible but not the epithelium. (F) At P0, Pax3 expression persists in the unfused palatal shelves (arrows) as well as in the tongue. Original magnification, ×6.3.
Persistent Pax3 expression impairs osteogenic differentiation.
In order to determine whether the cleft palate was due to an osteogenic defect, we examined palate mineralization at critical developmental time points. At E15.5, palatal shelves have normally lifted and fused at the midline, with epithelial to mesenchymal transition occurring at the fusion seam. By E15.5, mineralization at the periphery of the palate is normally visible (Figure 5A). Goldner’s trichrome–stained sections through the palates of E15.5 and E17.5 Pax3Cre/+, R26Pax3/Pax3 embryos revealed a distinct decrease in mineralization compared with control, with an absence of mineralization of the ossifying centers at the midline at E17.5 (Figure 5A). This decrease in mineralization was also evident at embryonic E14.5 and E16.5 (data not shown).
Figure 5. Persistent Pax3 expression impairs palate osteogenesis.
(A) Frontal sections through the palatal regions of E15.5 (top panels) and E17.5 (bottom panels) control (Pax3Cre/+, R26+/+), heterozygous (Pax3Cre/+, R26Pax3/+), and homozygous (Pax3Cre/+, R26Pax3/Pax3) littermates persistently expressing Pax3 are stained for mineralized bone (green) by Goldner’s trichrome. Pax3Cre/+, R26Pax3/Pax3 embryos have less mineralized bone in the palate compared with control (arrows). At E17.5, Pax3Cre/+, R26Pax3/Pax3 embryos lacked the medial mineralizing centers (arrows) seen in control (Pax3Cre/+, R26+/+) and heterozygotes (Pax3Cre/+, R26Pax3/+). Original magnification, ×2.5. (B) Von Kossa–stained ex vivo cultures of primary palate cells grown for 9 days in mineralizing medium. Control cells (left wells) formed mineralized nodules with BMP-2 treatment, but cells derived from Pax3Cre/+, R26Pax3/Pax3 mice (right wells) failed to form nodules and mineralize. Replicate wells are shown for each genotype. (C) Primary palate cell cultures stained for ALP activity. Control cells (left panel) cultured in osteogenic medium containing BMP-2 respond with robust expression of ALP, but cells from Pax3Cre/+, R26Pax3/Pax3 (right panel) mice are unable to respond to BMP-2 with upregulated ALP expression. Original magnification, ×16. (D) Immunofluorescent detection of Pax3 demonstrates persistent expression in cells from mutant mice (right panels) and the absence of Pax3 protein in control (Pax3Cre/+, R26+/+) cells. (E) Western blot analysis for Pax3 expression of primary palate cell culture lysates from control (Pax3Cre/+, R26+/+), heterozygous (Pax3Cre/+, R26Pax3/+), and homozygous (Pax3Cre/+, R26Pax3/Pax3) mice persistently expressing Pax3. Scale bar: 10 μm.
The decrease in palate mineralization suggested a defect in osteoblastogenesis. We isolated primary palate mesenchymal cells from E14.5 embryos and cultured them in the presence or absence of BMP-2 to determine their osteogenic potential. Primary palate cultures from control mice were able to form nodules and mineralize ex vivo, as shown by Von Kossa staining, when treated with BMP-2 (Figure 5B) but not in the absence of BMP-2 (data not shown). In contrast, BMP-treated primary palate cultures that persistently expressed Pax3 failed to form nodules and mineralize (Figure 5B). In order to determine whether the cause of the block in mineralization was due to a defect in osteoblast differentiation, we examined the expression of alkaline phosphatase (ALP), an early marker of osteoblastogenesis. Similar to our mineralization results, primary palate cultures from control mice responded robustly to BMP-2 treatment with high levels of ALP expression. However, palate cultures persistently expressing Pax3 failed to increase ALP levels in response to BMP-2 treatment (Figure 5C). Persistent expression of Pax3 in the primary palate cultures was confirmed by immunofluorescence (Figure 5D). Nearly all cells isolated in the primary palate cell preparation were positive for Pax3, indicating that the primary cultures were primarily mesenchymal cells derived from cranial neural crest of the Pax3-expressing linage. In contrast, cells isolated from control mice had no detectable Pax3, consistent with the previously observed downregulation of endogenous Pax3 at this time point. Western blot analysis confirmed that Pax3 was detectable in cells from mice persistently expressing Pax3, with lower levels in the heterozygous (Pax3Cre/+, R26Pax3/+) cells than in the homozygous (Pax3Cre/+, R26Pax3/Pax3) cells. No Pax3 was detected in cells from control littermates (Figure 5E).
Pax3-expressing cranial crest is resistant to BMP-induced signaling.
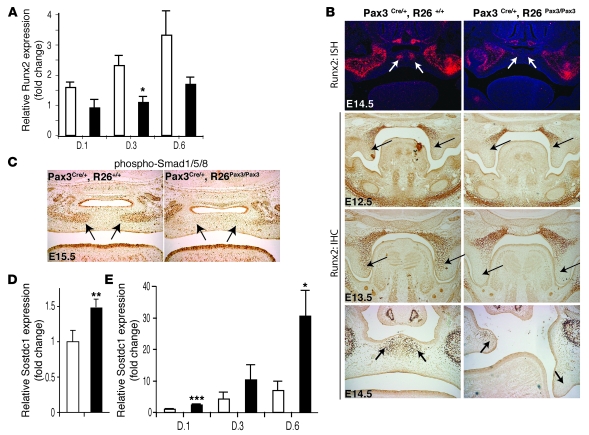
Despite abundant BMP expression in cranial regions, cranial crest normally fails to exhibit signs of osteogenic differentiation until Pax3 expression abates. Persistent expression of Pax3 further delays and impairs osteogenesis. Thus, we examined the effect of persistent Pax3 expression on early markers of BMP signaling and downstream targets. Runx2, a runt-domain transcription factor often described as the master regulator of osteoblast commitment and differentiation, is required for bone development (36–42) and functions downstream of BMPs (43, 44). Runx2-null mice die postnatally with respiratory failure due to a complete lack of mineralized bone (38). Overexpression of Runx2 in mesenchymal cell lines induces the expression of bone cell markers such as collagen type 1, ALP, and osterix, an osteoblast-specific protein (36, 38, 39, 42, 45). To examine the effect of Pax3 on Runx2 expression in palatal mesenchyme, quantitative PCR (QPCR) analysis was performed on RNA isolated from primary palate cells cultured for 1, 3, and 6 days in the presence of BMP-2. Cells persistently expressing Pax3 consistently expressed lower levels of Runx2 at all 3 time points (Figure 6A). In situ hybridization analysis of Runx2 expression levels demonstrated a decrease in Runx2 mRNA in the palates of Pax3Cre/+, R26Pax3/Pax3 mice at E14.5 compared with wild-type littermates, particularly in the ossifying centers at the midline (Figure 6B). Immunohistochemical analysis of E12.5, E13.5, and E14.5 palates using anti-Runx2 antibody staining confirmed the decrease in Runx2 levels in the palate, which was more pronounced at E13.5 and E14.5, when endogenous Pax3 levels were decreased (Figure 6B). The decrease in Runx2 expression was more pronounced in posterior areas of the developing palate (Supplemental Figure 3), consistent with our observation that the most penetrant aspects of the cleft palate phenotype are found in posterior regions. Similar observations of regional heterogeneity of gene expression in the developing palate have been described previously (46).
Figure 6. Persistent expression of Pax3 inhibits Runx2 expression and increases expression of the BMP-2 inhibitor Sostdc1.
(A) QPCR analysis of Runx2 expression in primary palate cell cultures at days 1, 3, and 6 of BMP–2 treatment (100 ng/ml). Control cells (white bars) express increasing levels of Runx2 relative to day 1 (D.1). Cells from embryos persistently expressing Pax3 (Pax3Cre/+, R26Pax3/Pax3; black bars), have significantly less Runx2 expression at all time points. (B) Runx2 mRNA levels in the palatal shelves (arrows) are decreased in the palatal region of E14.5 embryos compared with control littermates (left panels), as demonstrated by in situ hybridization with a probe for Runx2. Immunohistochemistry with anti-Runx2 antibody (bottom panels) also confirms that Runx2 protein is decreased in Pax3Cre/+, R26Pax3/Pax3 palates (right panels) compared with controls (left panels) in the palatal shelves (arrows) at E12.5, E13.5, and E14.5. Original magnification, ×2.5 (top 6 panels); ×6.3 (bottom 2 panels). (C) Immunohistochemistry on frontal sections of the palatal region of E15.5 embryos with an antibody against phosphorylated Smad1/5/8 demonstrates a decrease in Smad activation in the palates (arrows) of Pax3Cre/+, R26Pax3/Pax3 mice (right panel) compared with control littermates (left panel). Original magnification, ×6.3. (D) QPCR of RNA isolated from whole palatal shelves of E14.5 control (white bars) and Pax3Cre/+, R26Pax3/Pax3 littermates (black bars) shows a 50% increase in Sostsdc1 expression levels. (E) QPCR analysis of Sostdc1 expression in primary palate cell cultures at days 1, 3, and 6. Control cells (white bars) express low levels of Sostdc1, but cells derived from palates of littermates (Pax3Cre/+, R26Pax3/Pax3) persistently expressing Pax3 (black bars) have increasing levels of Sostdc1 up to 30-fold higher at D.6 than D.1 control levels. *P < 0.05; **P < 0.01; ***P < 0.001. Error bars represent SEM from 3 independent experiments, each performed in triplicate.
Resistance to BMP signaling would be expected to be associated with diminished activation of downstream components of the pathway, such as phosphorylation of the BMP-dependent Smads 1, 5, and 8. During normal palate development, phosphorylated Smad1/5/8 is first detected at approximately E13.5, coincident with downregulation of Pax3, and this is followed shortly thereafter by activation of Runx2 expression and osteogenic differentiation (data not shown). Immunohistological examination of sectioned E15.5 palatal shelves reveals a marked decrease of phosphorylated Smad1/5/8 in the regions of defective osteoblast differentiation and mineralization in the Pax3Cre/+, R26Pax3/Pax3 embryos compared with wild-type littermates (Figure 6C).
Upregulation of Sostdc1, an inhibitor of BMP signaling.
The data summarized above are consistent with diminished BMP responsiveness in cranial crest cells that express Pax3. We sought to identify the mechanism responsible for relative resistance of Pax3-expressing mesenchyme to BMP-mediated signals. Quantitative RT-PCR analysis (not shown) and in situ expression analysis of candidate components of the BMP signaling pathway, including both type 1 BMP receptors (BMPR-1a and BMPR-1b) and BMP-2, failed to reveal significant changes in Pax3Cre/+, R26Pax3/Pax3 embryos compared with controls (Supplemental Figure 4). However, microarray analysis of RNA isolated from dissected E14.5 palatal shelves of Pax3Cre/+, R26Pax3/Pax3 and control littermate embryos revealed a nearly 2-fold increase in expression of the secreted BMP inhibitor Sostdc1, also known as Ectodin. This was confirmed by QPCR, which revealed a 1.5-fold increase (Figure 6D). Furthermore, QPCR analysis of RNA from day 1, 3, and 6 primary palate cultures revealed a progressive increase in Sostdc1 produced by cells persistently expressing Pax3 when compared with controls (Figure 6E), while expression of the other BMP inhibitors Noggin, Tob, and Gremlin was not significantly altered (data not shown).
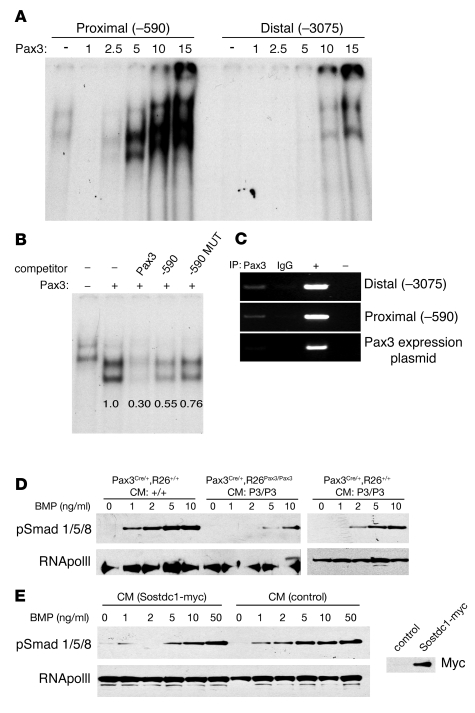
Reanalysis of our microarray data using modified bioinformatics algorithms was performed to identify potential regulatory elements found more commonly in up- or downregulated genes than in those that are unchanged. We evaluated 5 kb of genomic sequence upstream of the coding regions of the top 50 up- or downregulated genes and searched for any 10-bp nucleotide motifs that were overrepresented within noncoding genomic regions that were conserved between mouse and human sequences using the PhyME 1.2 program. We identified a motif with the consensus sequence AAAGACAGAA that occurred at a statistically significant higher frequency in regulated gene sequences than was predicted by chance; this sequence achieved the highest computational composite score according to an a priori weighting strategy. Interestingly, this consensus bears resemblance to the optimal Pax3 paired-domain–binding site previously described (5 of 9 bp matches; ref. 47). The highest scoring differentially regulated gene identified by this approach was Sostdc1, which contains at least 2 consensus sites within the 5-kb upstream region (–590 and –3075).
We empirically tested for Pax3 binding to the consensus Sostdc1 promoter motifs by electrophoretic mobility shift assays. Incubation of increasing amounts of nuclear extracts from Pax3-transfected 293T cells resulted in increasing amounts of bound (shifted) probe for both the proximal (–590) and distal (–3075) motifs, although significantly more extract was required to shift the distal motif probe (Figure 7A), suggesting lower affinity. Unlabeled proximal (–590) motif DNA effectively competes for binding, and unlabeled competitor DNA containing a previously validated Pax3 paired-domain–binding site (48) was somewhat more effective in competition assays than the –590 probe itself. A mutated proximal motif in which 4 residues within the binding element are altered is unable to effectively compete with labeled probe for binding to Pax3 (Figure 7B).
Figure 7. Pax3 binds the Sostdc1 promoter and decreases BMP-2 responsiveness.
(A) Electrophoretic mobility shift assay with radiolabeled probes for the proximal (–590) and distal (–3075) putative Pax3-binding motifs. Increasing amounts of nuclear extracts from Pax3-transfected 293T cells results in increased binding to both probes. Bands in first lane represent nonspecific binding. (B) Incubation with unlabeled competitor containing a Pax3-binding site (Pax3) or unlabeled proximal (–590) motif competitor reduces Pax3 binding to the proximal (–590) probe. A mutated proximal competitor (–590 Mut) competes poorly. Bands in the first lane represent nonspecific binding. Relative pixel densities of bands are indicated. (C) Immunoprecipitation of Sostdc1 promoter fragments containing either the distal or the proximal putative Pax3-binding motif are detected (lane 1, top and center panels) but not the Pax3 expression plasmid negative control (bottom panel). ChIP with control IgG1 demonstrates the specificity of the immunoprecipitation. IgG (lane 2) serves as a negative control. 293T lysate (+, lane 3) was used as a positive control. (D) Western blot to detect phosphorylated Smad1/5/8 or RNA polymerase II as loading control in nuclear extracts of primary palate cells treated with increasing doses of BMP-2. Lanes 1–5, control Pax3Cre/+, R26+/+ cells with control-conditioned medium (CM:+/+); lanes 6–10, Pax3Cre/+, R26Pax3/Pax3 cells with Pax3Cre/+, R26Pax3/Pax3–conditioned medium (CM:P3/P3); right panel, control cells with Pax3Cre/+, R26Pax3/Pax3–conditioned medium. (E) Western blot of nuclear extracts from wild-type primary palate cells treated with conditioned medium from control or myc-Sostdc1–transfected 293T cells and increasing doses of BMP-2. RIght panel demonstrates presence of myc-tagged Sostdc1 in conditioned medium from transfected cells.
Pax3 can also interact with the Sostdc1 upstream genomic region as determined by ChIP assays. ChIP analysis demonstrates that Pax3 interacts with Sostdc1 promoter fragments that contain either the proximal (–590) motif or the distal (–3075) motif (Figure 7C). Control experiments with nonspecific IgG1 immunoglobulin do not precipitate these fragments, and control DNA fragments are not precipitated with the Pax3-specific antibody, demonstrating the specificity of the interaction (Figure 7C).
These results suggested that Pax3 could regulate expression of Sostdc1 in cranial mesenchyme. Hence, we asked whether this mechanism could account for BMP resistance and osteogenic defects identified in Pax3Cre/+, R26Pax3/Pax3 mice. BMP-2 induces phosphorylation of Smad1/5/8 in wild-type primary palate cultures; however, palate cultures persistently expressing Pax3 show impaired Smad1/5/8 phosphorylation in response to BMP-2 (Figure 7D). Importantly, conditioned medium harvested from the cultures persistently expressing Pax3 is sufficient to mimic the blunted response to BMP-2 treatment in control cells, suggesting that a secreted factor from the Pax3Cre/+, R26Pax3/Pax3 cells is indeed responsible for this effect (Figure 7D). Conditioned medium from wild-type cultures did not display this activity. Wild-type primary palate cultures treated with conditioned medium from 293T cells transfected with myc-tagged Sostdc1 show a similar blunted response of Smad1/5/8 phosphorylation in response to BMP-2. This effect was not seen when cells were treated with conditioned medium from control-transfected cells (Figure 7E). Thus, we conclude that sustained expression of Pax3 in neural crest-derived palatal mesenchyme results in decreased BMP signaling and Smad activation associated with increased levels of the secreted BMP inhibitor Sostdc1 as well as defective osteogenic differentiation.
Discussion
We have examined the necessity for inactivation of Pax3 during neural crest differentiation. Our knockin strategy to maintain expression of Pax3 in Pax3-expressing neural crest cells beyond the time at which endogenous expression would have normally abated has revealed what we believe is a novel role for Pax3 in craniofacial skeletogenesis and osteoblast differentiation and maturation. During early patterning, undifferentiated cranial crest must resist differentiation cues in order to migrate and proliferate appropriately. Likewise, adult stem cells must employ mechanisms to remain undifferentiated until appropriate environmental signals are recognized. Our results suggest that 1 mechanism by which neural crest cells can resist osteogenic differentiation is by secretion of the soluble BMP inhibitor Sostdc1. Thus, our data show that persistent Pax3 expression causes cranioskeletal defects including cleft or shortened palates that result in neonatal lethality. Persistent expression of Sostdc1 results in diminished BMP responsiveness, as seen by decreased phosphorylated Smad1/5/8 in the palatal mesenchyme. This is associated with diminished osteogenesis and decreased expression of Runx2, a “master regulator” of osteoblast commitment and maturation that is required for bone mineralization (36, 38, 41).
Cleft palate is a common birth defect that occurs in 0.1%–0.2% of live births worldwide (49). The formation of the secondary palate requires a series of complex and highly coordinated events, beginning with the elongation of the palatal shelves at about E11 and continuing to the elevation and fusion of these shelves above the tongue by E14.5, separating the oral and nasal cavities. Mineralization of the palate occurs via intramembranous ossification in which condensations of mesenchymal cells directly differentiate into osteoblasts (46, 50). Several factors have been implicated in palatogenesis including sonic hedgehog, FGFs, Snail, TGF-β, and BMP receptors, although these reports do not provide mechanistic explanations for failure of palatal shelves to lift (49, 51, 52). However, we did not observe obvious alterations in the expression of these genes (Supplemental Figures 4–6 and data not shown). Although subtle changes in expression that are below the detection threshold of in situ hybridization cannot be ruled out, QPCR analysis of BMPR-1a, -1b, and -2 did not reveal any difference in transcript levels.
Persistent expression of Pax3 from the Rosa26 locus under the control of the Pax3Cre knockin recapitulates proper spatial expression domains for Pax3 (27), and expression levels appeared similar to endogenous levels. Consistent with this, expression from the Rosa26 locus is sufficient to largely rescue embryonic development in mice otherwise null for Pax3. Cleft palate was evident when Pax3 was persistently expressed in mice homozygous for the R26Pax3 allele, regardless of whether 0, 1, or 2 alleles of endogenous Pax3 were present. Also, palate defects were not evident until time points when endogenous Pax3 expression was normally decaying. These observations indicate that the defects in palatogenesis that we describe are due to persistent Pax3 expression rather than to altered levels of Pax3 in cranial crest during early time points when Pax3 is normally expressed. Further, our analyses of the Pax3-expressing cells in the developing palatal shelves indicate that migration and survival of these cells are not affected and thus not likely to be a cause of the defects in palatal development.
We hypothesize that expression of Pax3 may function in part to retain mesenchymal progenitor cells in an undifferentiated state, and abatement of Pax3 expression is necessary for normal progression of osteoblast differentiation. Upregulation of the BMP-inhibitor Sostdc1 resulting from direct transcriptional regulation by Pax3 may result in a delay or block in osteoblastogenesis by blocking BMP and Smad signaling–induced commitment and/or differentiation by mechanisms that include inhibition of Runx2 expression. This model is consistent with the role of Pax3 in melanocyte precursors, where it functions to activate expression of Mitf, while simultaneously inhibiting Mitf from activating downstream genes and melanocyte differentiation (9). Thus, adult melanocyte stem cells that continue to express Pax3 are prevented from melanocytic differentiation. In cranial crest, Pax3 is likely to regulate Sostdc1 expression by working in concert with other transcription factors, as we failed to detect significant modulation of Sostdc1 reporter constructs by Pax3 in luciferase transcription assays (data not shown). Although the present work focuses on the osteogenic defect in the developing palate, additional defects in neural crest–derived bones were observed. The alisphenoid, pterygoid processes of the basisphenoid and tympanic bones, which were all affected, are derived from neural crest (30–33), suggesting a broad effect of persistent Pax3 expression on neural crest–derived osteogenic tissues.
Surprisingly, we did not observe gross defects in many neural crest–derived tissues in mice with persistent expression of Pax3. For example, the outflow tract of the heart and the smooth muscle layer of the aortic arch develop properly despite maintained Pax3 expression (Figure 1D). However, defects in adult tissues derived from Pax3 lineage precursors could not be evaluated due to neonatal lethality of the mice persistently expressing Pax3. Several factors may account for the lack of additional embryonic defects. It is possible that persistent Pax3 activity is well tolerated in some tissues. Also, it is likely that Pax3 activity is regulated by multiple mechanisms (ref. 53 and our unpublished observations) in addition to transcriptional regulation and that Pax3 protein expression does not directly reflect transcriptional activity. Absence of required cofactors or the presence of inhibitory factors might result in a dampening of the functional activity of persistent Pax3 expression in some tissues. In melanoblasts, for example, active Wnt signaling appears to result in displacement of Pax3 from some enhancers (9). In the case of skeletal muscle development, although Pax3Cre/+, R26Pax3/Pax3 pups have normal limb musculature at birth, neonatal lethality precludes an examination of possible adult muscle phenotypes and satellite cell function during maintenance and regeneration of skeletal muscle. Although Pax3Cre/+, R26Pax3/+ heterozygous adults do not have an obvious defect in muscle growth and maintenance (data not shown), it is clear that 1 copy of the R26Pax3 allele is not sufficient to produce a dramatic phenotype.
In summary, we have utilized an in vivo genetic approach to address the question of whether inactivation of a developmentally critical transcription factor is necessary for proper fate determination and tissue development. Our results suggest that temporal regulation of Pax3 expression during embryogenesis is critical for cranial neural crest development and suggest that both activation and inactivation of transcriptional regulators should be the focus of future analysis.
Methods
Knockin mouse generation and genotyping.
The Rosa26-Pax3 knockin construct was generated by inserting the Pax3 cDNA (GenBank accession number NM_008781) from pCMV-Pax3dhfr (20) into pBigT (41). Subsequent cloning steps to produce the targeting vector and targeted R1 ES cells were essentially as described (41). Rosa26-Pax3 mice were genotyped in a duplex PCR reaction with the following primers: R26F2, 5′-AAAGTCGCTCTGAGTTGTTAT-3′; R523, 5′-GGAGCGGGAGAAATGGATATG-3′; and R1295, 5′-GCGAAGAGTTTGTCCTCAACC-3′, which produced a wild-type band of 584 bp and a mutant band of 250 bp. Pax3Cre/+ mice were genotyped as previously described (27). All animal studies were approved by the University of Pennsylvania Institutional Animal Care and Use Committee.
Immunoblot.
Total cell lysates, nuclear extracts, or whole-embryo lysates were prepared and run on 4%–12% SDS-PAGE gels. Protein concentration was measured by BCA assay (Pierce Biotechnology) and equal amounts of total protein were loaded. Protein gels were transferred onto nitrocellulose membranes and blotted with monoclonal Pax3 antibody (University of Iowa Developmental Studies Hybridoma Bank).
Immunohistochemistry and in situ hybridization.
Immunohistochemistry was performed using monoclonal Pax3 antibody, polyclonal PEBP2aA (M-70) antibody (Santa Cruz Biotechnology Inc.) against Runx2, and pSmad1/5/8 antibody no. 9511 (Cell Signaling Technology). Runx2 in situ hybridization (54) was performed using a 300-bp EcoR1/XbaI fragment of the Runx2 5′UTR (36). Intensities of control and experimental images were always adjusted to the same degree using Adobe Photoshop CS3.
Skeleton and primary palate culture staining.
Alizarin red/Alcian blue staining was performed on newborn pups after removing skin and internal organs. The carcasses were fixed in 90% ethanol, then placed in 0.1 mg/ml Alcian blue 8GX in 20% (v/v) glacial acetic acid/80% (v/v) ethanol for 7 days. After rehydration, the samples were placed in fresh 1% (w/v) potassium hydroxide (KOH) for 24 hours. After the samples became translucent, they were placed in 0.2 (w/v) mg/ml alizarin red S in freshly prepared 1% KOH for 2–3 days. The stained skeletons were further cleared in fresh 1% (w/v) KOH and then dehydrated in glycerol for imaging and storage. Goldner’s trichrome staining was performed for 5 minutes in ponceau/acid fuchsin (0.75 g ponceau 2R [ponceau xylidine] [P2395-25G; Sigma-Aldrich], 0.25 g acid fuchsin, 0.1 g azophloxine [11640-25G; Sigma-Aldrich]), 5 minutes in orange G (20 g orange G [861286-25G; Sigma-Aldrich], 40 g phosphomolybdic acid, 1 l with distilled water), and 5 minutes in light green (2 g light green SF [L1886-25G; Sigma-Aldrich], 2 ml glacial acetic acid, to 1 l with distilled water) with acetic acid rinses in between each step. Von Kossa and ALP staining were performed as described (55).
Primary palate cultures.
Paired palatal shelves were dissected from E14.5 embryos and placed in cold PBS with 2% Fungizone (Invitrogen) in individual wells of a 24-well dish. The following steps were performed under sterile conditions in a tissue culture hood: the PBS was aspirated and replaced with 750 μl of 0.25% trypsin (Invitrogen) and incubated at 4°C for 10 minutes followed by 37°C for 5 minutes. Palates were then dissociated by trituration in 0.25% trypsin with a 1-ml pipette. Trypsinization was halted by addition of 80 μl FBS, and the dissociated cells were seeded in 1 well of a 6-well dish for each embryo. Cells were cultured in α-MEM, 10% FBS, and 1% Fungizone (Invitrogen) for 2–3 days until nearly confluent and then expanded into a 10-cm dish. Cells were seeded for Von Kossa and ALP staining in a 12-well dish at 100,000 cells/well in α-MEM, 10% FBS, and 1% penicillin/streptomycin. At confluence, cells were treated with 100 ng/ml BMP-2 (R&D Systems). For BMP treatment, cells were seeded in 6-cm culture dishes at approximately 400,000 cells/dish and grown until confluent, about 2–3 days. At confluence, cells were starved overnight in 1.5 ml total volume starvation medium: Opti-MEM (Invitrogen), 1% FBS, and 1% penicillin/streptomycin. The next day, conditioned medium was collected and spun at 2000 g for 4 minutes to remove dead cells and debris. BMP was added at the stated concentrations, and then the treated conditioned medium was replaced on the cells. Cells were incubated with BMP-treated conditioned medium for 30 minutes at 37°C, after which nuclear extracts were prepared.
Sostdc1-conditioned medium preparation.
293T cells were seeded in 10-cm dishes and transfected overnight with 5 μg of myc-tagged Sostdc1 or control vector using FuGENE 6 reagent as described (Roche Applied Science). The day after transfection, medium was replaced with 5 ml of Opti-MEM (Invitrogen), 1% FBS, and 1% penicillin/streptomycin. Conditioned medium was collected 48 and 72 hours after transfection, and cellular debris was pelleted at 2000 g for 4 minutes prior to use.
QPCR.
First-strand cDNA was prepared using either Superscript Reverse Transcriptase III (Invitrogen) or High Capacity cDNA Synthesis (Applied Biosystems) according to the product guides. Runx2 (Mm0501578_m1) was assayed with TaqMan probes and master mixes (Applied Biosystems) normalized to Gapdh (Mm9999915_g1). Sostdc1 was assayed using SYBR green master mixes (Applied Biosystems) and the following primer set: Sostdc1F: 5′–CCATTGCTCTTTCCTCACTT-3′, Sostdc1R: 5′-GATGGAAACTACAGGATGCC 3′. SYBR green assays were normalized to Gapdh using the following primers: GapdhF: 5′-TGTCTTCACCACCATGGAGAAGG-3′, GapdhR: 5′-GTGGATGCAGGGATGATGTTCTG-3′. Assays were performed in triplicate, and the averages of at least 3 independent experiments are shown. Statistical analysis was performed using 1-tailed Student’s t test relative to the control sample for each condition. P < 0.5 was considered significant.
Microarray analysis.
Microarray analysis was performed on 8 pairs of dissected palatal shelves from E14.5 Pax3Cre/+, R26+/+ and Pax3Cre/+, R26Pax3/Pax3 embryos collected from 2 litters. Palates were homogenized in 1 ml of Trizol (Invitrogen), and RNA was prepared using RNeasy columns (QIAGEN) and eluted in 30 μl RNase-free water. Purified RNA was submitted to the Penn Microarray Facility ( http://www.med.upenn.edu/microarr) for analysis on Mouse Affymetrix MOE430 v2.0 chips. Statistical analyses of gene expression were performed using BioConductor open-source software packages in R ( http://www.bioconductor.org). Cells from 8 mice spread across 2 litters were used, and fluorescence intensity measures were normalized by robust multiarray analysis (56). To identify differentially expressed transcripts we used the limma package (57), which adopts a Bayesian approach. The identification of differentially expressed transcripts controlled for effects of variations between mouse litters. R-scripts for these analyses are available in Supplemental Methods, and R-script output is provided in Supplemental Table 2. Microarray data will be available at GEO Profiles, NCBI.
Genome sequence extraction.
All Affymetrix spots with nominally significant differential expression (P < 0.01) were ranked in descending order according to their expression fold-change (wild type versus mutated). The top 50 probes were selected for subsequent analysis. The Affymetrix IDs for these 50 spots were then converted into Ensembl IDs ( www.ensembl.org), and 5000 bp sequences upstream and downstream of their associated mouse genes were extracted using the BioMart database ( www.ensembl.org). We retained only those genes with orthologs in the human genome, of which there were 30. Upstream and downstream sequences from the coding region of the human orthologs were also extracted.
Motif-finding analysis.
Motif-finding analysis was conducted using the PhyME 1.2 package ( http://edsc.rockefeller.edu/cgi-bin/phyme/download.pl). Parameter settings were as follows: 2 species, 30 sequences, 10-bp motif, flat phylogeny, 3 motifs, and 0.80 threshold instance. Only the motif with the highest expectation maximization score from the output file was used for this analysis. Genes annotated by NCBI and containing this motif in aligned upstream regions with a threshold instance greater than 0.88 were taken as putative downstream targets of Pax3. Out of this set of genes, the ones containing this motif in more than 2 aligned region were ranked as the most likely targets.
ChIP.
ChIP was performed using 293T cells cotransfected with Pax3 expression vector and 2 pGL3PR constructs containing fragments of the Sostdc1 promoter cloned from genomic DNA by restriction digestion of BAC DNA (RP23-98E22; Invitrogen) with EcoR1 (distal: –3180 to –2128) or PCR with the following primers: forward, 5′-CAGCACTTACTTAGGCAGCAGACTC-3′; reverse, 5′-GCTTGGCGTTCATTCAGGTG-3′ (proximal: –1017 to –108). Immunoprecipitation of cross-linked complexes was performed as described (58), with the following modifications: nuclear lysates were measured by protein assay, and 10 μg of lysates were precleared with 50 μl of protein A/G–conjugated agarose beads (Santa Cruz Biotechnology Inc.) in 1 ml total volume mRIPA (with 350 mM NaCl) for 1 hour at 4°C with rotation. Following complex immunoprecipitation, elution, and DNA purification, samples were analyzed by PCR for the presence of Sostdc1 promoter fragments using the following primers: distal motif (–3075) forward: 5′-AGCACTGACCAATGTTTTGA-3′, reverse: 5′-TTTCCTTCAAGTCCATGTTG-3′; proximal motif (–590) forward: 5′-CTCATTACACCCCCCAACCC-3′; reverse: 5′-TACCACTTGCTCCTCAGCCAGGTCCTCTCC-3′; Pax3 forward: 5′-AGACCGACTATGCTCTCTCC-3′, reverse: 5′-TTTGTCCATACTGCCCATAC-3′. PCR was performed with annealing at 56°C for 25 cycles. Products were run on 2% agarose gels.
Electrophoretic mobility shift assay.
Nuclear extracts of 293T cells transfected with control vector or Pax3 expression vector were preincubated with an excess of cold competitor oligos or Pax3 antibody (1 μg) for 30 minutes in a total volume of 20 μl as previously described (59). Sostdc1 probes were made by annealing the following oligos: proximal (–590) forward: 5′-GTCTTAAGAGACAAAACATAT-3′, reverse: 5′-GATATGTTTTGTCTCTTAAGA-3′; distal (–3075) forward: 5′-GCAGAGGAAGACAGTAGTCAC-3′, reverse: 5′-GGTGACTACTGTCTTCCTCTG-3′. They were then labeled with 32P-αdCTP using the MegaPrime labeling kit (GE Healthcare). Approximately 30,000 cpm of probe was added to each reaction. Cold competitor was made by annealing the following oligos: Pax3 forward: 5′-GCATGCAGAGTCTGTGCTTCCAACCACCATGTCACACTGCCCATG-3′, reverse: 5′-GCATGGGCAGTGTGACATGGTGGTTGGAAGCACAGACTCTGCATG-3′; –590 Mut forward: 5′-GTCTTACCAGACAAGGCATAT-3′, reverse: 5′-GATATGCCTTGTCTGGTAAGA-3′. Samples were run on 6% nondenaturing Tris-Borate-EDTA (TBE) gels, dried under vacuum, and exposed to film at –80°C.
Supplementary Material
Acknowledgments
We would like to thank Nicole Antonucci, Lan Cheng, and Maozhen Zhang for excellent animal care and technical support. This work was supported by funding from the University of Pennsylvania Training Program in Cardiovascular Biology and Medicine and a training grant from the University of Pennsylvania Medical Center Institute for Medicine and Engineering (to M. Wu), grants from the Burroughs Wellcome Fund and the James S. McDonnell Foundation (to J.B. Plotkin), and grants from the NIH (RO1 HL61475 to J. Epstein) and the W.W. Smith Endowed Chair at the University of Pennsylvania (to J.A. Epstein).
Footnotes
Nonstandard abbreviations used: ALP, alkaline phosphatase; BMP, bone morphogenetic protein; Pax3, paired box 3; QPCR, quantitative PCR; Sostdc1, sclerostin domain–containing 1.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 118:2076–2087 (2008). doi:10.1172/JCI33715
References
- 1.Choi J., et al. MyoD converts primary dermal fibroblasts, chondroblasts, smooth muscle, and retinal pigmented epithelial cells into striated mononucleated myoblasts and multinucleated myotubes. Proc. Natl. Acad. Sci. U. S. A. 1990;87:7988–7992. doi: 10.1073/pnas.87.20.7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudnicki M.A., et al. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-V. [DOI] [PubMed] [Google Scholar]
- 3.Tajbakhsh S., et al. Gene targeting the myf-5 locus with nlacZ reveals expression of this myogenic factor in mature skeletal muscle fibres as well as early embryonic muscle. Dev. Dyn. 1996;206:291–300. doi: 10.1002/(SICI)1097-0177(199607)206:3<291::AID-AJA6>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 4.Weintraub H., et al. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc. Natl. Acad. Sci. U. S. A. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 6.Goulding M.D., Chalepakis G., Deutsch U., Erselius J.R., Gruss P. Pax-3, a novel murine DNA binding protein expressed during early neurogenesis. EMBO J. 1991;10:1135–1147. doi: 10.1002/j.1460-2075.1991.tb08054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chi N., Epstein J.A. Getting your Pax straight: Pax proteins in development and disease. Trends Genet. 2002;18:41–47. doi: 10.1016/S0168-9525(01)02594-X. [DOI] [PubMed] [Google Scholar]
- 8.Stoller J.Z., Epstein J.A. Cardiac neural crest. Semin Cell Dev. Biol. 2005;16:704–715. doi: 10.1016/j.semcdb.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Lang D., et al. Pax3 functions at a nodal point in melanocyte stem cell differentiation. Nature. 2005;433:884–887. doi: 10.1038/nature03292. [DOI] [PubMed] [Google Scholar]
- 10.Tajbakhsh S., Rocancourt D., Cossu G., Buckingham M. Redefining the genetic hierarchies controlling skeletal myogenesis: Pax-3 and Myf-5 act upstream of MyoD. Cell. 1997;89:127–138. doi: 10.1016/S0092-8674(00)80189-0. [DOI] [PubMed] [Google Scholar]
- 11.Conboy I.M., Rando T.A. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell. 2002;3:397–409. doi: 10.1016/S1534-5807(02)00254-X. [DOI] [PubMed] [Google Scholar]
- 12.Montarras D., et al. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- 13.Bennicelli J.L., Advani S., Schafer B.W., Barr F.G. PAX3 and PAX7 exhibit conserved cis-acting transcription repression domains and utilize a common gain of function mechanism in alveolar rhabdomyosarcoma. Oncogene. 1999;18:4348–4356. doi: 10.1038/sj.onc.1202812. [DOI] [PubMed] [Google Scholar]
- 14.Xia S.J., Pressey J.G., Barr F.G. Molecular pathogenesis of rhabdomyosarcoma. Cancer Biol. Ther. 2002;1:97–104. doi: 10.4161/cbt.51. [DOI] [PubMed] [Google Scholar]
- 15.Kozmik Z. Pax genes in eye development and evolution. Curr. Opin. Genet. Dev. 2005;15:430–438. doi: 10.1016/j.gde.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 16.Marquardt T., et al. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/S0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 17.Del Rio-Tsonis K., Washabaugh C.H., Tsonis P.A. Expression of pax-6 during urodele eye development and lens regeneration. Proc. Natl. Acad. Sci. U. S. A. 1995;92:5092–5096. doi: 10.1073/pnas.92.11.5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pennisi E. Evolution of developmental diversity. Evo-devo devotees eye ocular origins and more. Science. 2002;296:1010–1011. doi: 10.1126/science.296.5570.1010. [DOI] [PubMed] [Google Scholar]
- 19.Melchers F. B cell development and its deregulation to transformed states at the pre-B cell receptor-expressing pre-BII cell stage. Curr. Top. Microbiol. Immunol. 2005;294:1–17. doi: 10.1007/3-540-29933-5_1. [DOI] [PubMed] [Google Scholar]
- 20.Li J., Liu K.C., Jin F., Lu M.M., Epstein J.A. Transgenic rescue of congenital heart disease and spina bifida in Splotch mice. Development. 1999;126:2495–2503. doi: 10.1242/dev.126.11.2495. [DOI] [PubMed] [Google Scholar]
- 21.Lang D., Epstein J.A. Sox10 and Pax3 physically interact to mediate activation of a conserved c-RET enhancer. Hum. Mol. Genet. 2003;12:937–945. doi: 10.1093/hmg/ddg107. [DOI] [PubMed] [Google Scholar]
- 22.Potterf S.B., Furumura M., Dunn K.J., Arnheiter H., Pavan W.J. Transcription factor hierarchy in Waardenburg syndrome: regulation of MITF expression by SOX10 and PAX3. Hum. Genet. 2000;107:1–6. doi: 10.1007/s004390050001. [DOI] [PubMed] [Google Scholar]
- 23.Watanabe A., Takeda K., Ploplis B., Tachibana M. Epistatic relationship between Waardenburg syndrome genes MITF and PAX3. Nat. Genet. 1998;18:283–286. doi: 10.1038/ng0398-283. [DOI] [PubMed] [Google Scholar]
- 24.Mayanil C.S., et al. Regulation of murine TGFbeta2 by Pax3 during early embryonic development. J. Biol. Chem. 2006;281:24544–24552. doi: 10.1074/jbc.M512449200. [DOI] [PubMed] [Google Scholar]
- 25.Mansouri A., Gruss P. Pax3 and Pax7 are expressed in commissural neurons and restrict ventral neuronal identity in the spinal cord. Mech. Dev. 1998;78:171–178. doi: 10.1016/S0925-4773(98)00168-3. [DOI] [PubMed] [Google Scholar]
- 26.Zambrowicz B.P., et al. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc. Natl. Acad. Sci. U. S. A. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Engleka K.A., et al. Insertion of Cre into the Pax3 locus creates a new allele of Splotch and identifies unexpected Pax3 derivatives. Dev. Biol. 2005;280:396–406. doi: 10.1016/j.ydbio.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Kioussi C., Gross M.K., Gruss P. Pax3: a paired domain gene as a regulator in PNS myelination. Neuron. 1995;15:553–562. doi: 10.1016/0896-6273(95)90144-2. [DOI] [PubMed] [Google Scholar]
- 29.Chai Y., et al. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- 30.Chai Y., Maxson R.E., Jr. Recent advances in craniofacial morphogenesis. Dev. Dyn. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- 31.Couly G.F., Coltey P.M., Le Douarin N.M. The triple origin of skull in higher vertebrates: a study in quail-chick chimeras. Development. 1993;117:409–429. doi: 10.1242/dev.117.2.409. [DOI] [PubMed] [Google Scholar]
- 32.Noden D.M., Schneider R.A. Neural crest cells and the community of plan for craniofacial development: historical debates and current perspectives. Adv. Exp. Med. Biol. 2006;589:1–23. doi: 10.1007/978-0-387-46954-6_1. [DOI] [PubMed] [Google Scholar]
- 33.Noden D.M., Trainor P.A. Relations and interactions between cranial mesoderm and neural crest populations. J. Anat. 2005;207:575–601. doi: 10.1111/j.1469-7580.2005.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Creuzet S., Vincent C., Couly G. Neural crest derivatives in ocular and periocular structures. Int. J. Dev. Biol. 2005;49:161–171. doi: 10.1387/ijdb.041937sc. [DOI] [PubMed] [Google Scholar]
- 35.Danielian P.S., et al. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 1998;8:1323–1326. doi: 10.1016/S0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 36.Ducy P., et al. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev. 1999;13:1025–1036. doi: 10.1101/gad.13.8.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ducy P., Zhang R., Geoffroy V., Ridall A.L., Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/S0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 38.Komori T., et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/S0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 39.Lian J.B., Stein G.S. Runx2/Cbfa1: a multifunctional regulator of bone formation. Curr. Pharm. Des. 2003;9:2677–2685. doi: 10.2174/1381612033453659. [DOI] [PubMed] [Google Scholar]
- 40.Liu W., et al. Overexpression of Cbfa1 in osteoblasts inhibits osteoblast maturation and causes osteopenia with multiple fractures. J. Cell Biol. 2001;155:157–166. doi: 10.1083/jcb.200105052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otto F., et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/S0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 42.Xiao Z.S., Hinson T.K., Quarles L.D. Cbfa1 isoform overexpression upregulates osteocalcin gene expression in non-osteoblastic and pre-osteoblastic cells. J. Cell. Biochem. 1999;74:596–605. doi: 10.1002/(SICI)1097-4644(19990915)74:4<596::AID-JCB9>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 43.Lee K.S., et al. Runx2 is a common target of transforming growth factor beta1 and bone morphogenetic protein 2, and cooperation between Runx2 and Smad5 induces osteoblast-specific gene expression in the pluripotent mesenchymal precursor cell line C2C12. Mol. Cell. Biol. 2000;20:8783–8792. doi: 10.1128/MCB.20.23.8783-8792.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee M.H., et al. Transient upregulation of CBFA1 in response to bone morphogenetic protein-2 and transforming growth factor beta1 in C2C12 myogenic cells coincides with suppression of the myogenic phenotype but is not sufficient for osteoblast differentiation. J. Cell. Biochem. 1999;73:114–125. doi: 10.1002/(SICI)1097-4644(19990401)73:1<114::AID-JCB13>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 45.Nakashima K., et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/S0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 46.Okano J., Suzuki S., Shiota K. Regional heterogeneity in the developing palate: morphological and molecular evidence for normal and abnormal palatogenesis. Congenit. Anom. (Kyoto) 2006;46:49–54. doi: 10.1111/j.1741-4520.2006.00103.x. [DOI] [PubMed] [Google Scholar]
- 47.Epstein J.A., Shapiro D.N., Cheng J., Lam P.Y., Maas R.L. Pax3 modulates expression of the c-Met receptor during limb muscle development. Proc. Natl. Acad. Sci. U. S. A. 1996;93:4213–4218. doi: 10.1073/pnas.93.9.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lang D., et al. Pax3 is required for enteric ganglia formation and functions with Sox10 to modulate expression of c-ret. J. Clin. Invest. 2000;106:963–971. doi: 10.1172/JCI10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gritli-Linde A. Molecular control of secondary palate development. Dev. Biol. 2007;301:309–326. doi: 10.1016/j.ydbio.2006.07.042. [DOI] [PubMed] [Google Scholar]
- 50.Kerrigan J.J., Mansell J.P., Sengupta A., Brown N., Sandy J.R. Palatogenesis and potential mechanisms for clefting. J. R. Coll. Surg. Edinb. 2000;45:351–358. [PubMed] [Google Scholar]
- 51.Murray S.A., Oram K.F., Gridley T. Multiple functions of Snail family genes during palate development in mice. Development. 2007;134:1789–1797. doi: 10.1242/dev.02837. [DOI] [PubMed] [Google Scholar]
- 52.Rice R., et al. Disruption of Fgf10/Fgfr2b-coordinated epithelial-mesenchymal interactions causes cleft palate. J. Clin. Invest. 2004;113:1692–1700. doi: 10.1172/JCI20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boutet S.C., Disatnik M.H., Chan L.S., Iori K., Rando T.A. Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell. 2007;130:349–362. doi: 10.1016/j.cell.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 54.Wawersik S., Epstein J.A. Gene expression analysis by in situ hybridization. Radioactive probes. Methods Mol. Biol. 2000;137:87–96. doi: 10.1385/1-59259-066-7:87. [DOI] [PubMed] [Google Scholar]
- 55.Sabatakos G., et al. Overexpression of DeltaFosB transcription factor(s) increases bone formation and inhibits adipogenesis. Nat. Med. 2000;6:985–990. doi: 10.1038/79683. [DOI] [PubMed] [Google Scholar]
- 56.Irizarry R.A., et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Smyth, G.K. 2005. Limma: linear models for microarray data. InBioinformatics and computational biology solutions using R and bioconductor. V.C.R. Gentleman, S. Dudoit, R. Irizarry, and W. Huber, editors. Springer. New York, New York, USA. 397–420. [Google Scholar]
- 58.Wiebe P.O., et al. Ptf1a binds to and activates area III, a highly conserved region of the Pdx1 promoter that mediates early pancreas-wide Pdx1 expression. Mol. Cell. Biol. 2007;27:4093–4104. doi: 10.1128/MCB.01978-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Epstein J.A., Lam P., Jepeal L., Maas R.L., Shapiro D.N. Pax3 inhibits myogenic differentiation of cultured myoblast cells. J. Biol. Chem. 1995;270:11719–11722. doi: 10.1074/jbc.270.20.11719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.