Abstract
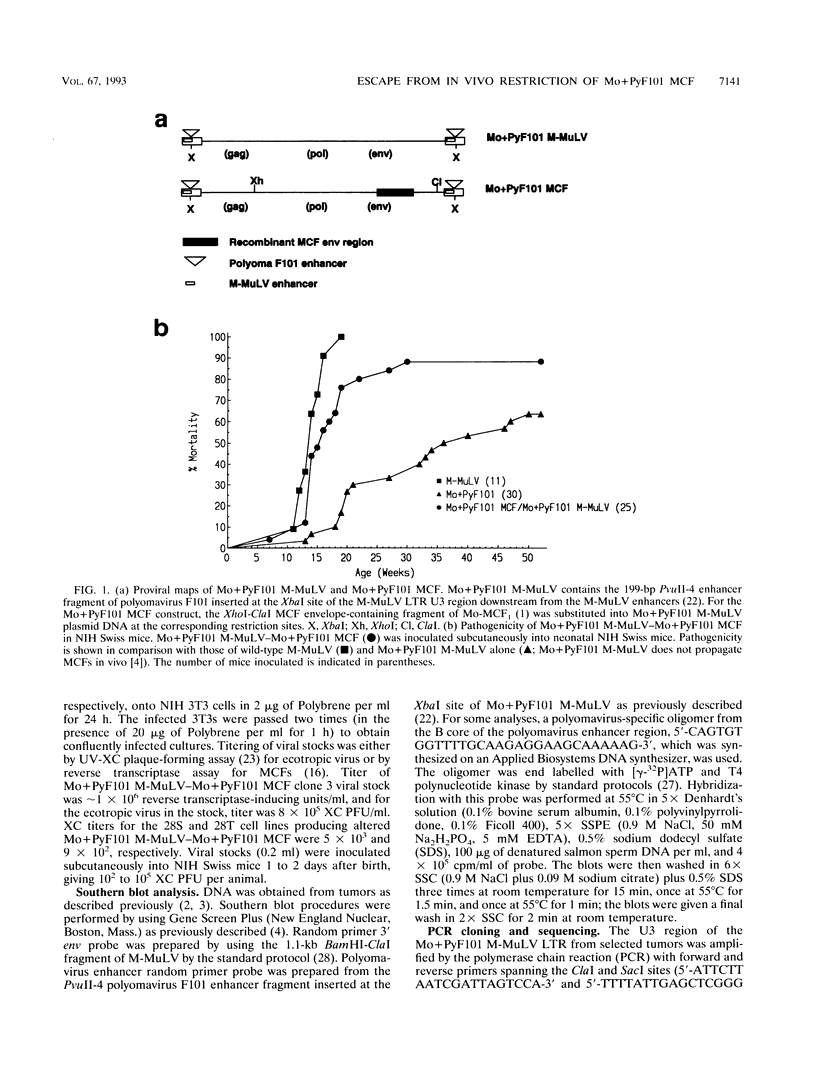
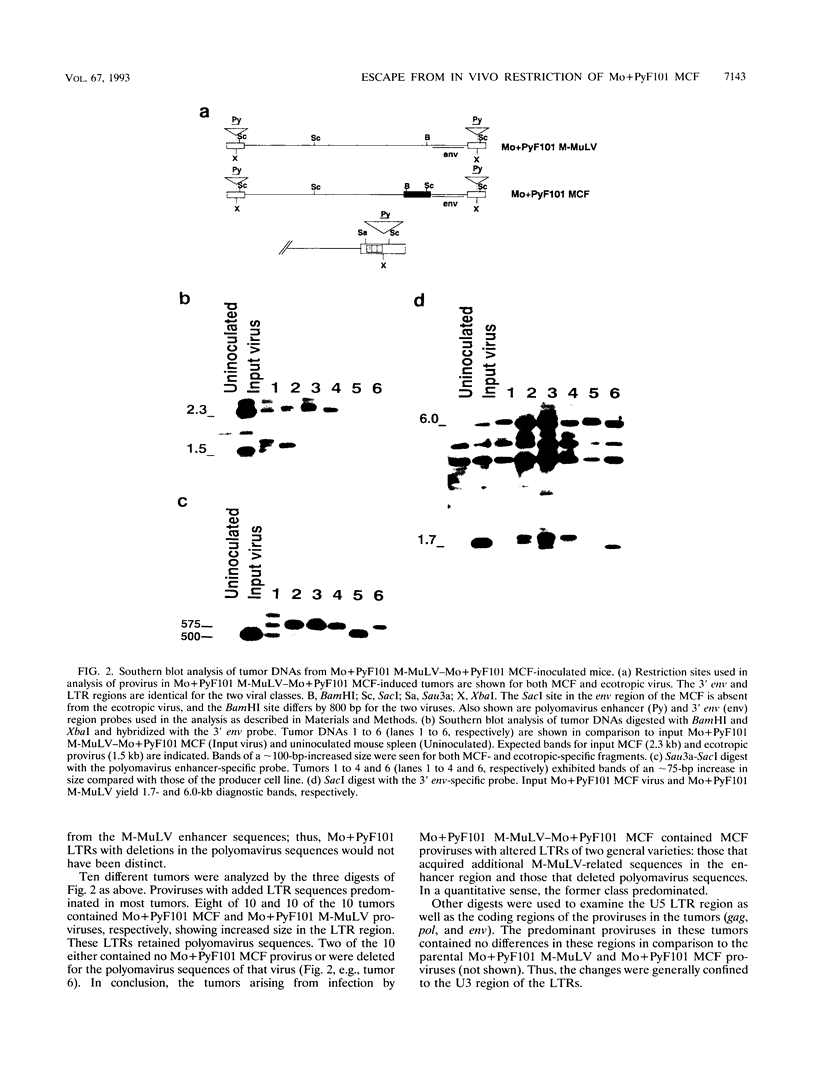
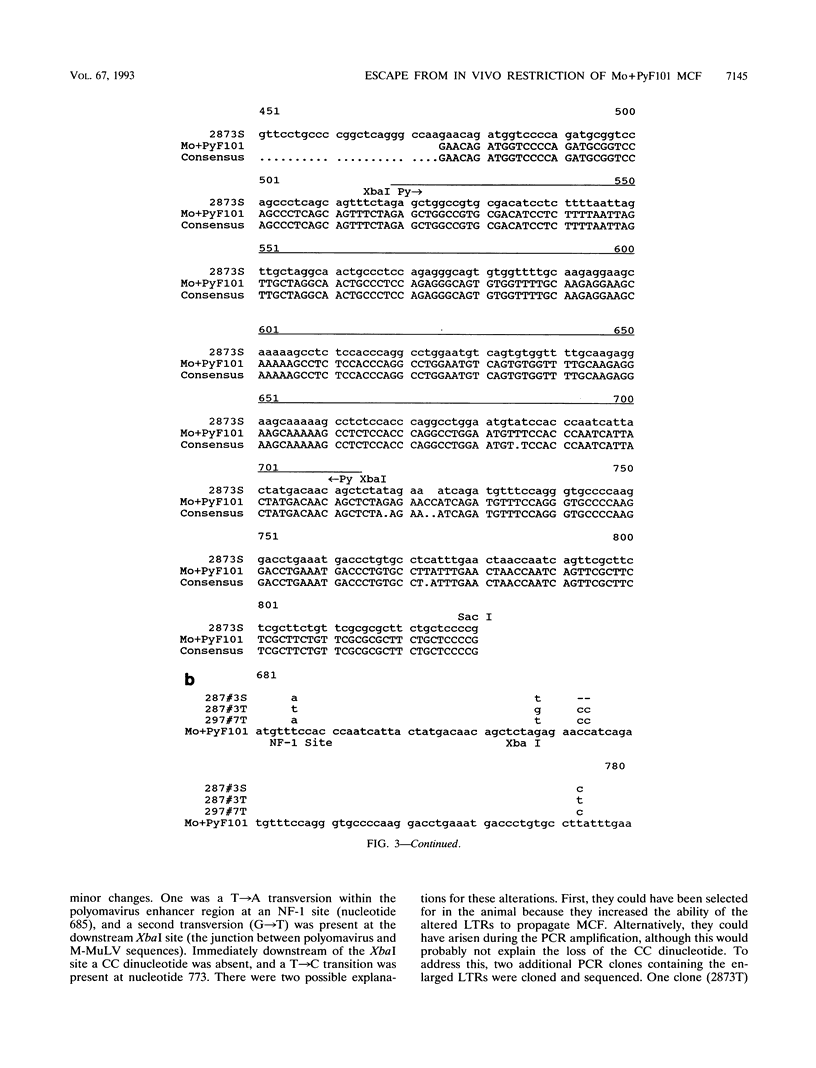
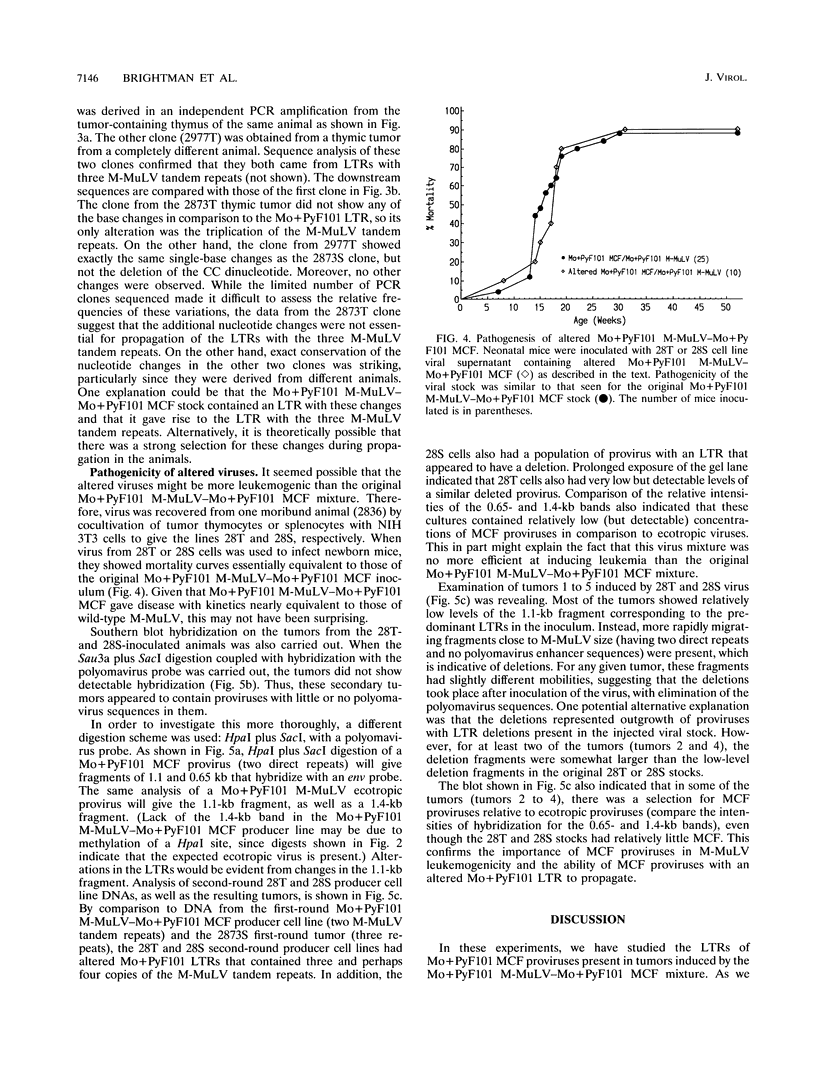
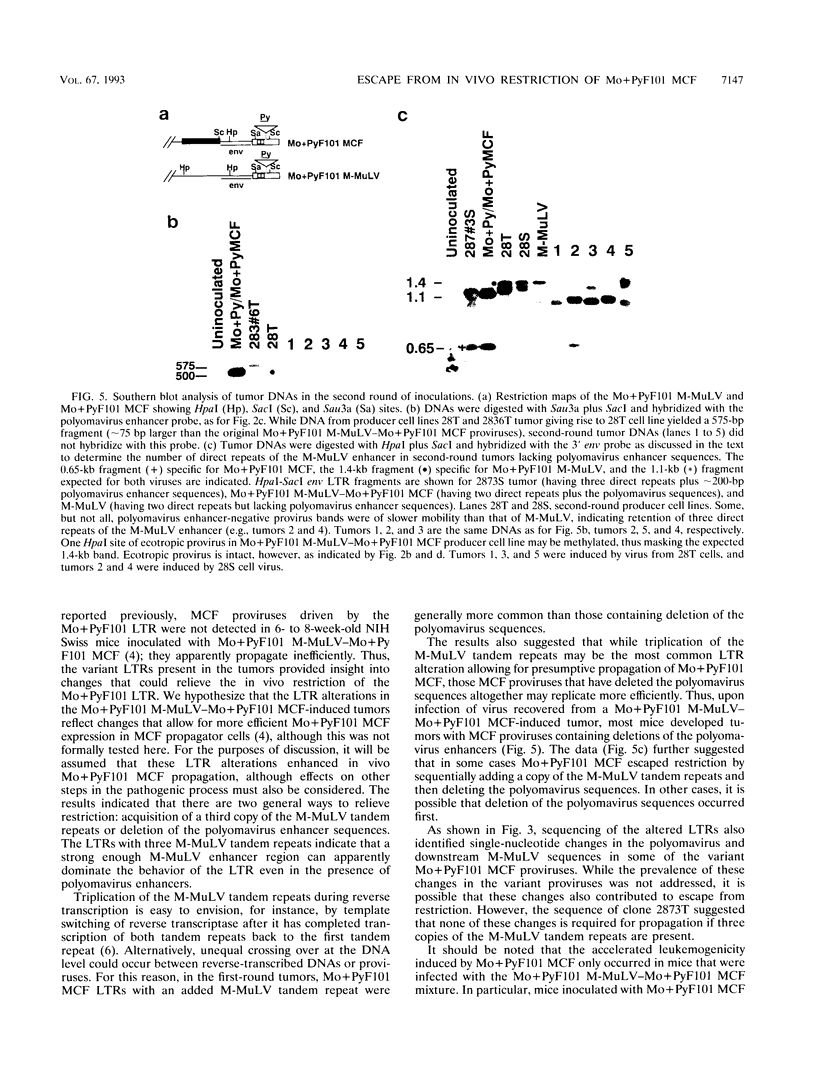
Mo+PyF101 M-MuLV is a variant Moloney murine leukemia virus containing polyomavirus F101 enhancers inserted just downstream from the M-MuLV enhancers in the long terminal repeat (LTR). The protein coding sequences for this virus are identical to those of M-MuLV. Mo+PyF101 M-MuLV induces T-cell disease with a much lower incidence and longer latency than wild-type M-MuLV. We have previously shown that Mo+PyF101 M-MuLV is defective in preleukemic events induced by wild-type M-MuLV, including splenic hematopoietic hyperplasia, bone marrow depletion, and generation of recombinant mink cell focus-inducing viruses (MCFs). We also showed that an M-MCF virus driven by the Mo+PyF101 LTR is infectious in vitro but does not propagate in mice. However, in these experiments, when a pseudotypic mixture of Mo+PyF101 M-MuLV and Mo+PyF101 MCF was inoculated into newborn NIH Swiss mice, they died of T-cell leukemia at times almost equivalent to those induced by wild-type M-MuLV. Tumor DNAs from Mo+PyF101 M-MuLV-Mo+PyF101 MCF-inoculated mice were examined by Southern blot analysis. The predominant forms of Mo+PyF101 MCF proviruses in these tumors contained added sequences in the U3 region of the LTR. The U3 regions of representative tumor-derived variant Mo+PyF101 MCFs were cloned by polymerase chain reaction amplification, and sequencing indicated that they had acquired an additional copy of the M-MuLV 75-bp tandem repeat in the enhancer region. NIH 3T3 cell lines infected with altered viruses were obtained from representative Mo+PyF101 M-MuLV-Mo+PyF101 MCF-induced tumors, and mice were inoculated with the recovered viruses. Leukemogenicity was approximately equivalent to that in the original Mo+PyF101 M-MuLV-Mo+PyF101 MCF viral stock. Southern blot analysis on the resulting tumors now predominantly revealed loss of the polyomavirus sequences. These results suggest that the suppressive effects of the PyF101 sequences on M-MuLV-induced disease and potentially on MCF propagation were overcome in two ways: by triplication of the M-MuLV direct repeats and by loss of the polyomavirus sequences.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bosselman R. A., van Straaten F., Van Beveren C., Verma I. M., Vogt M. Analysis of the env gene of a molecularly cloned and biologically active Moloney mink cell focus-forming proviral DNA. J Virol. 1982 Oct;44(1):19–31. doi: 10.1128/jvi.44.1.19-31.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brightman B. K., Chandy K. G., Spencer R. H., Fan H. A T lymphoid cell line responds to a thymic stromal cell line by expression of Thy-1 and CD4. J Immunol. 1989 Nov 1;143(9):2775–2782. [PubMed] [Google Scholar]
- Brightman B. K., Chandy K. G., Spencer R. H., Gupta S., Pattengale P. K., Fan H. Characterization of lymphoid tumors induced by a recombinant murine retrovirus carrying the avian v-myc oncogene. Identification of novel (B-lymphoid) tumors in the thymus. J Immunol. 1988 Oct 15;141(8):2844–2854. [PubMed] [Google Scholar]
- Brightman B. K., Rein A., Trepp D. J., Fan H. An enhancer variant of Moloney murine leukemia virus defective in leukemogenesis does not generate detectable mink cell focus-inducing virus in vivo. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2264–2268. doi: 10.1073/pnas.88.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloyd M. W., Hartley J. W., Rowe W. P. Lymphomagenicity of recombinant mink cell focus-inducing murine leukemia viruses. J Exp Med. 1980 Mar 1;151(3):542–552. doi: 10.1084/jem.151.3.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuypers H. T., Selten G. C., Zijlstra M., de Goede R. E., Melief C. J., Berns A. J. Tumor progression in murine leukemia virus-induced T-cell lymphomas: monitoring clonal selections with viral and cellular probes. J Virol. 1986 Oct;60(1):230–241. doi: 10.1128/jvi.60.1.230-241.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuypers H. T., Selten G., Quint W., Zijlstra M., Maandag E. R., Boelens W., van Wezenbeek P., Melief C., Berns A. Murine leukemia virus-induced T-cell lymphomagenesis: integration of proviruses in a distinct chromosomal region. Cell. 1984 May;37(1):141–150. doi: 10.1016/0092-8674(84)90309-x. [DOI] [PubMed] [Google Scholar]
- Davis B. R., Brightman B. K., Chandy K. G., Fan H. Characterization of a preleukemic state induced by Moloney murine leukemia virus: evidence for two infection events during leukemogenesis. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4875–4879. doi: 10.1073/pnas.84.14.4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B. R., Chandy K. G., Brightman B. K., Gupta S., Fan H. Effects of nonleukemogenic and wild-type Moloney murine leukemia virus on lymphoid cells in vivo: identification of a preleukemic shift in thymocyte subpopulations. J Virol. 1986 Nov;60(2):423–430. doi: 10.1128/jvi.60.2.423-430.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B., Linney E., Fan H. Suppression of leukaemia virus pathogenicity by polyoma virus enhancers. Nature. 1985 Apr 11;314(6011):550–553. doi: 10.1038/314550a0. [DOI] [PubMed] [Google Scholar]
- Evans L. H., Cloyd M. W. Friend and Moloney murine leukemia viruses specifically recombine with different endogenous retroviral sequences to generate mink cell focus-forming viruses. Proc Natl Acad Sci U S A. 1985 Jan;82(2):459–463. doi: 10.1073/pnas.82.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans L. H., Morrey J. D. Tissue-specific replication of Friend and Moloney murine leukemia viruses in infected mice. J Virol. 1987 May;61(5):1350–1357. doi: 10.1128/jvi.61.5.1350-1357.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Chute H., Chao E., Pattengale P. K. Leukemogenicity of Moloney murine leukemia viruses carrying polyoma enhancer sequences in the long terminal repeat is dependent on the nature of the inserted polyoma sequences. Virology. 1988 Sep;166(1):58–65. doi: 10.1016/0042-6822(88)90146-8. [DOI] [PubMed] [Google Scholar]
- Fan H., Jaenisch R., MacIsaac P. Low-multiplicity infection of Moloney murine leukemia virus in mouse cells: effect on number of viral DNA copies and virus production in producer cells. J Virol. 1978 Dec;28(3):802–809. doi: 10.1128/jvi.28.3.802-809.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura F. K., Deininger P. L., Friedmann T., Linney E. Mutation near the polyoma DNA replication origin permits productive infection of F9 embryonal carcinoma cells. Cell. 1981 Mar;23(3):809–814. doi: 10.1016/0092-8674(81)90445-1. [DOI] [PubMed] [Google Scholar]
- Hedrick S. M., Cohen D. I., Nielsen E. A., Davis M. M. Isolation of cDNA clones encoding T cell-specific membrane-associated proteins. Nature. 1984 Mar 8;308(5955):149–153. doi: 10.1038/308149a0. [DOI] [PubMed] [Google Scholar]
- Holland C. A., Thomas C. Y., Chattopadhyay S. K., Koehne C., O'Donnell P. V. Influence of enhancer sequences on thymotropism and leukemogenicity of mink cell focus-forming viruses. J Virol. 1989 Mar;63(3):1284–1292. doi: 10.1128/jvi.63.3.1284-1292.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. X., Fan H. Bone marrow depletion by 89Sr complements a preleukemic defect in a long terminal repeat variant of Moloney murine leukemia virus. J Virol. 1991 Aug;65(8):4442–4448. doi: 10.1128/jvi.65.8.4442-4448.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. X., Fan H. Combined infection by Moloney murine leukemia virus and a mink cell focus-forming virus recombinant induces cytopathic effects in fibroblasts or in long-term bone marrow cultures from preleukemic mice. J Virol. 1990 Aug;64(8):3701–3711. doi: 10.1128/jvi.64.8.3701-3711.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linney E., Davis B., Overhauser J., Chao E., Fan H. Non-function of a Moloney murine leukaemia virus regulatory sequence in F9 embryonal carcinoma cells. 1984 Mar 29-Apr 4Nature. 308(5958):470–472. doi: 10.1038/308470a0. [DOI] [PubMed] [Google Scholar]
- Rowe W. P., Pugh W. E., Hartley J. W. Plaque assay techniques for murine leukemia viruses. Virology. 1970 Dec;42(4):1136–1139. doi: 10.1016/0042-6822(70)90362-4. [DOI] [PubMed] [Google Scholar]
- Selten G., Cuypers H. T., Zijlstra M., Melief C., Berns A. Involvement of c-myc in MuLV-induced T cell lymphomas in mice: frequency and mechanisms of activation. EMBO J. 1984 Dec 20;3(13):3215–3222. doi: 10.1002/j.1460-2075.1984.tb02281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Stoye J. P., Coffin J. M. The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J Virol. 1987 Sep;61(9):2659–2669. doi: 10.1128/jvi.61.9.2659-2669.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve L., Rassart E., Jolicoeur P., Graham M., Adams J. M. Proviral integration site Mis-1 in rat thymomas corresponds to the pvt-1 translocation breakpoint in murine plasmacytomas. Mol Cell Biol. 1986 May;6(5):1834–1837. doi: 10.1128/mcb.6.5.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]