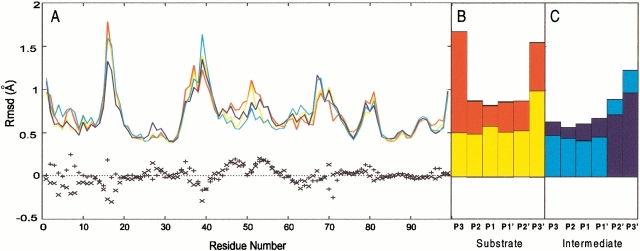
Fig. 4.
Average root mean square deviation (rmsd) calculated on each residue of the protein (A), peptide substrate (SUB) (B), gem-diol intermediate (INT) (C) relative to the average molecular dynamics structure: Wild type/SUB complex (red); M46I/SUB complex (yellow); wild type/INT complex (blue); M46I/INT complex (cyan). The difference between the value calculated for the wild type and the M46I enzyme is reported as `+' for the human immunodeficiency virus type 1 protease (HIV-1 PR)/SUB complexes and as `x' for the HIV-1 PR/INT complexes.