Figure 2.
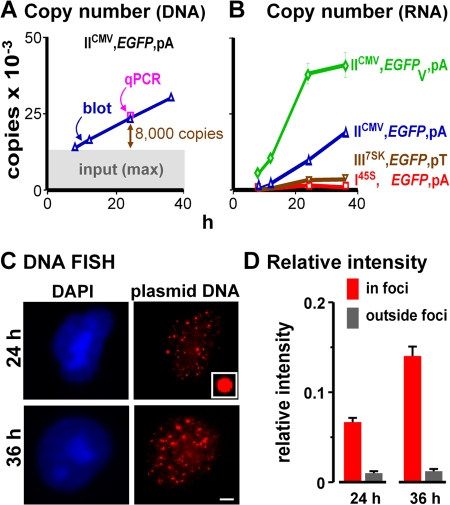
Replicating minichromosomes are concentrated in few foci. Cells were cotransfected with IICMV,DsRed,pA (so transfection efficiencies could be monitored by FACS using DsRed fluorescence) and another encoding EGFP as indicated; after 8 h, cells were replated (to wash away input) and regrown for 4, 16, and 28 h. (A) The DNA copy number of IICMV,EGFP,pA per transfected cell—determined by reference to known amounts of pure plasmid DNA (by blotting using a “Hirt” extract and qPCR using total DNA)—increases above the maximum possible background due to input (upper limit of gray area). Numbers (24 h) obtained in analogous experiments for I45S,EGFP,pA, III7SK,EGFP,pT, and IICMV,EGFPV,pA were similar (i.e., 23,000 ± 2,000, 18,000 ± 3,000, and 21,000 ± 3,000, respectively; one-way ANOVA, P > 0.05). (B) Promoters and introns affect the RNA copy number per transfected cell (determined using qRT-PCR by reference to known amounts of pure RNA). (C) DNA FISH shows that minichromosomes are concentrated in ∼20 foci. (inset) Fluorescent bead used for normalization. Bar, 2.5 μm. (D) Mean intensities (+SD) of pixels in or outside foci in 100 images like those in C were normalized relative to the beads; most signal is in the foci, and this increases with time.