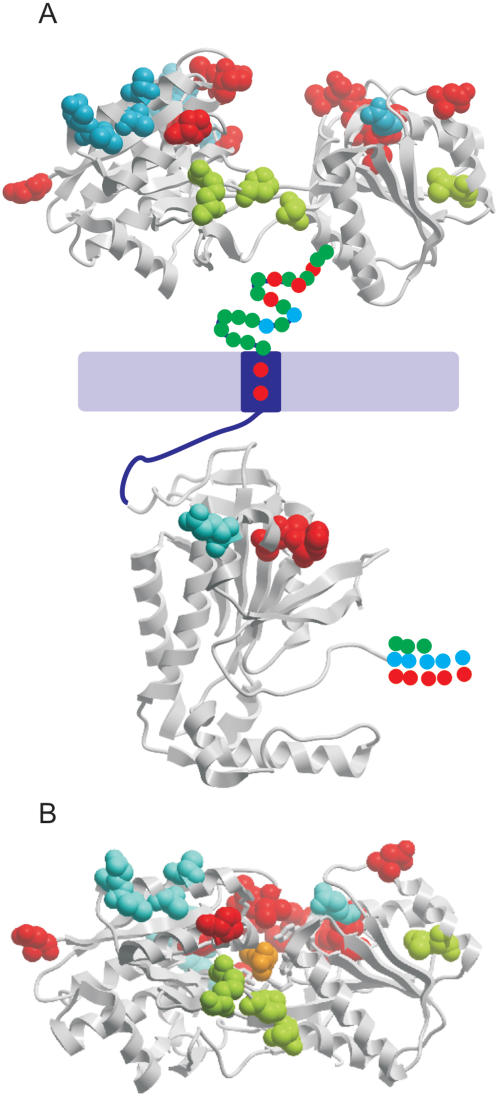
Figure 3. Codon-specific analysis of receptor-type adenylate cyclase genes.
Homologous extracellular codons with predicted ω>1 are mapped to the tertiary structure of E. coli leucine-binding protein (PDB 2LBP [28]) or L-leucine-binding protein with leucine bound (PDB 1USK [56]). Intracellular codons that are predicted to be under positive selection are mapped to the tertiary structure of T. brucei receptor-type adenylate cyclise (PDB 1FX2 [25]). A) Hypothetical representation of the intact Trypanosome adenylate cyclase molecule. The extracellular region is composed of an N-terminal region homologous to 2LBP and a region of unknown function. A single transmembrane region links to the conserved C-terminal catalytic domain. Positively selected sites (posterior probability ≥0.95) homologous to crystal structures are shown as space filled residues. Red, CPG 21943524. Blue, 20115358. Green, 19651441. Positively selected sites in regions not homologous to a known crystal structure are shown as coloured circles. B) Position of positively selected sites following conformation change of extracellular binding region upon binding to leucine (coloured orange). Orientation is as for part A.