Abstract
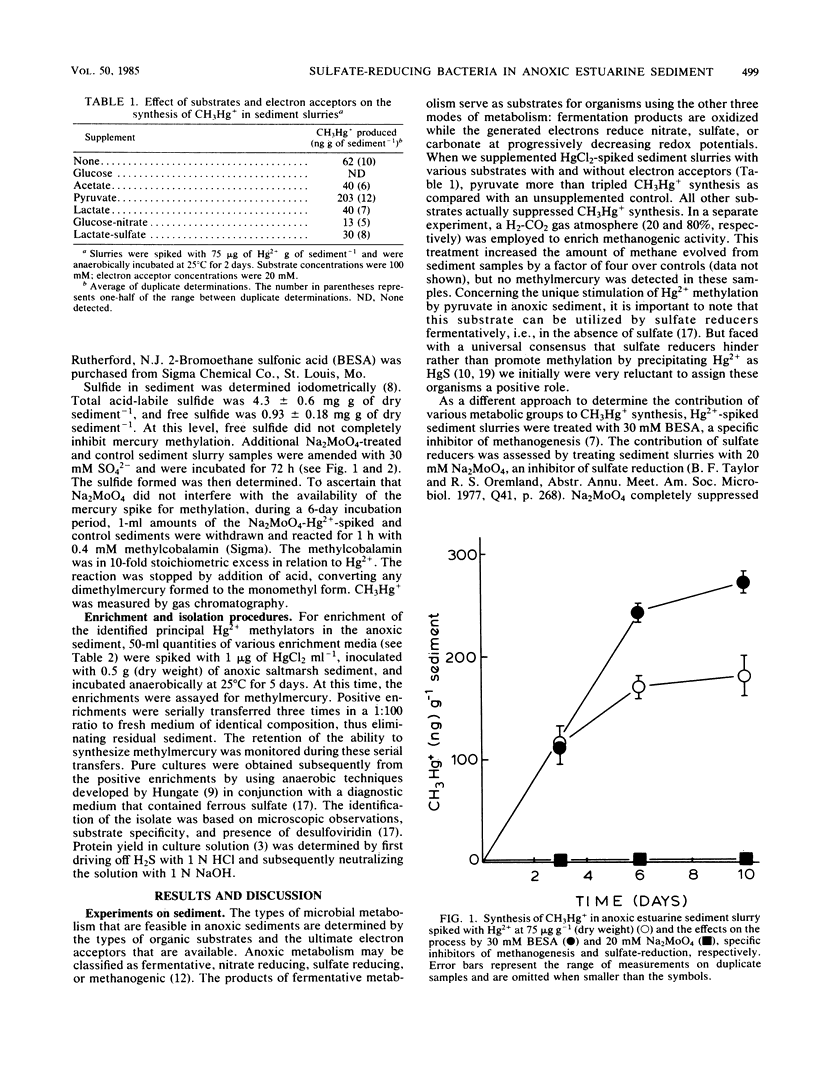
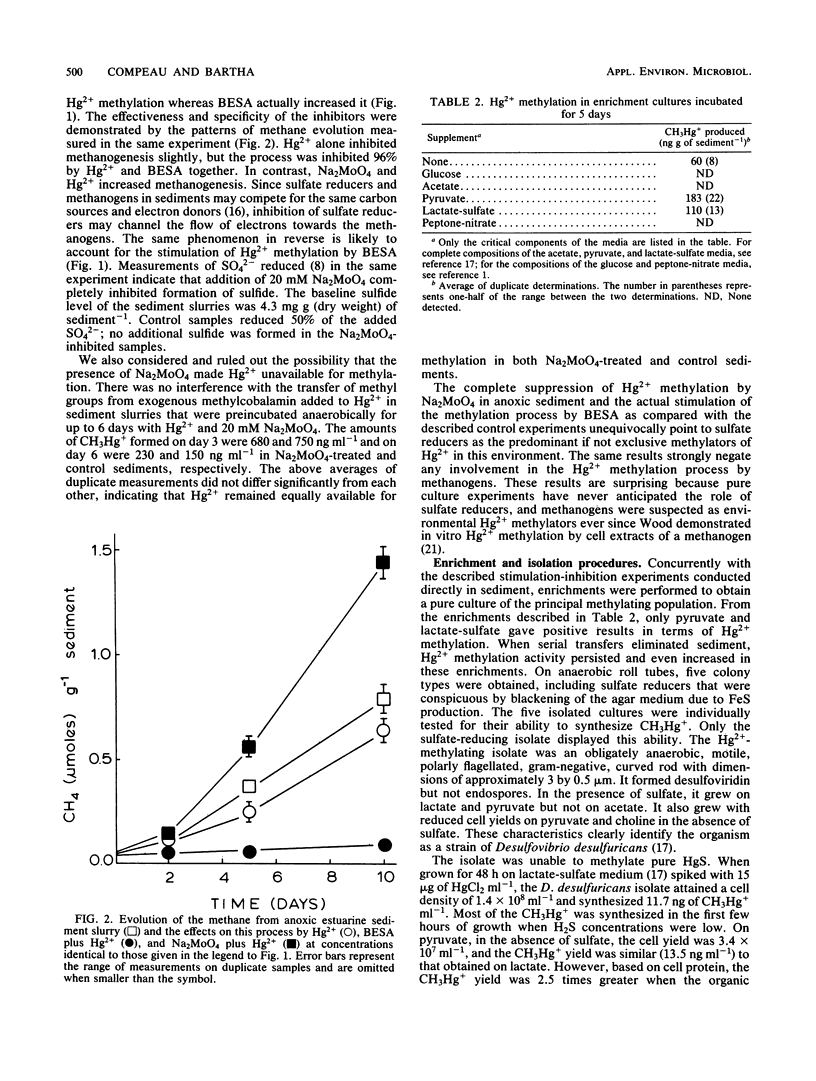
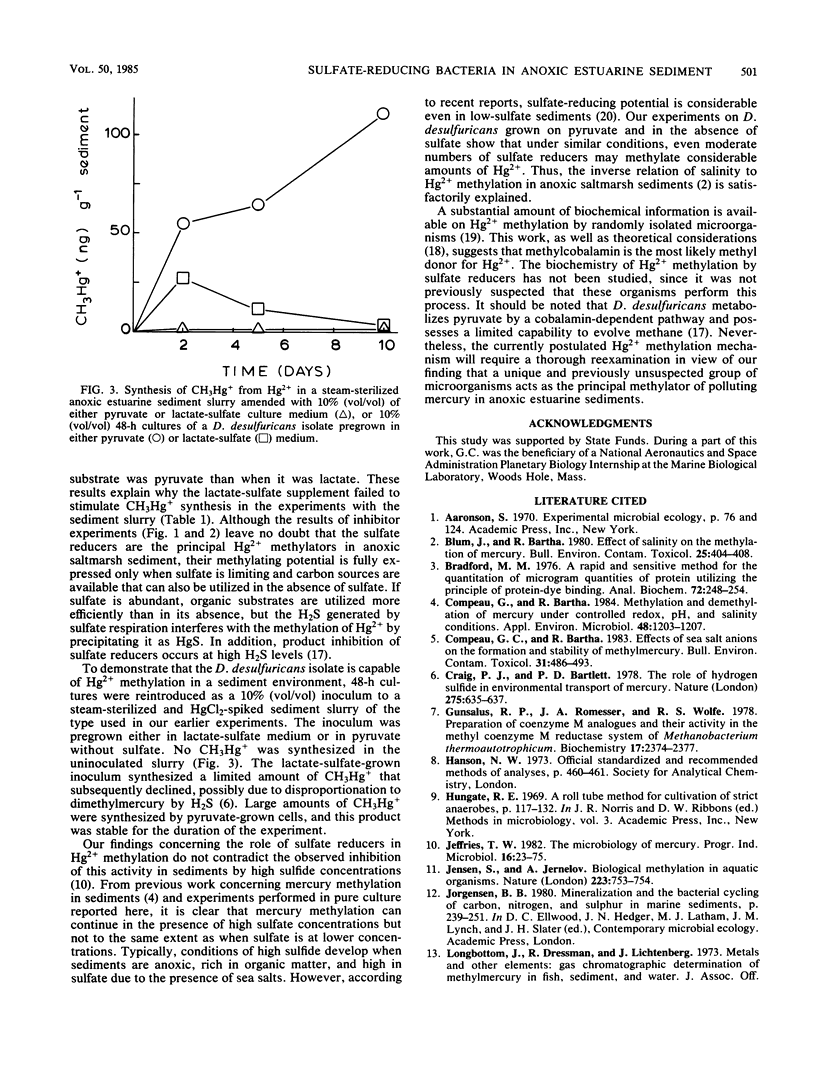
Substrate-electron acceptor combinations and specific metabolic inhibitors were applied to anoxic saltmarsh sediment spiked with mercuric ions (Hg2+) in an effort to identify, by a direct approach, the microorganisms responsible for the synthesis of hazardous monomethylmercury. 2-Bromoethane sulfonate (30 mM), a specific inhibitor of methanogens, increased monomethylmercury synthesis, whereas sodium molybdate (20 mM), a specific inhibitor of sulfate reducers, decreased Hg2+ methylation by more than 95%. Anaerobic enrichment and isolation procedures yielded a Desulfovibrio desulfuricans culture that vigorously methylated Hg2+ in culture solution and also in samples of presterilized sediment. The Hg2+ methylation activity of sulfate reducers is fully expressed only when sulfate is limiting and fermentable organic substrates are available. To date, sulfate reducers have not been suspected of Hg2+ methylation. Identification of these bacteria as the principal methylators of Hg2+ in anoxic sediments raises questions about the environmental relevance of previous pure culture-based methylation work.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blum J. E., Bartha R. Effect of salinity on methylation of mercury. Bull Environ Contam Toxicol. 1980 Sep;25(3):404–408. doi: 10.1007/BF01985546. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Compeau G., Bartha R. Effects of sea salt anions on the formation and stability of methylmercury. Bull Environ Contam Toxicol. 1983 Oct;31(4):486–493. doi: 10.1007/BF01622282. [DOI] [PubMed] [Google Scholar]
- Compeau G., Bartha R. Methylation and demethylation of mercury under controlled redox, pH and salinity conditions. Appl Environ Microbiol. 1984 Dec;48(6):1203–1207. doi: 10.1128/aem.48.6.1203-1207.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus R. P., Romesser J. A., Wolfe R. S. Preparation of coenzyme M analogues and their activity in the methyl coenzyme M reductase system of Methanobacterium thermoautotrophicum. Biochemistry. 1978 Jun 13;17(12):2374–2377. doi: 10.1021/bi00605a019. [DOI] [PubMed] [Google Scholar]
- Jensen S., Jernelöv A. Biological methylation of mercury in aquatic organisms. Nature. 1969 Aug 16;223(5207):753–754. doi: 10.1038/223753a0. [DOI] [PubMed] [Google Scholar]
- Ridley W. P., Dizikes L. J., Wood J. M. Biomethylation of toxic elements in the environment. Science. 1977 Jul 22;197(4301):329–332. doi: 10.1126/science.877556. [DOI] [PubMed] [Google Scholar]
- Robinson J. B., Tuovinen O. H. Mechanisms of microbial resistance and detoxification of mercury and organomercury compounds: physiological, biochemical, and genetic analyses. Microbiol Rev. 1984 Jun;48(2):95–124. doi: 10.1128/mr.48.2.95-124.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. L., Klug M. J. Reduction of sulfur compounds in the sediments of a eutrophic lake basin. Appl Environ Microbiol. 1981 May;41(5):1230–1237. doi: 10.1128/aem.41.5.1230-1237.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J. M., Kennedy F. S., Rosen C. G. Synthesis of methyl-mercury compounds by extracts of a methanogenic bacterium. Nature. 1968 Oct 12;220(5163):173–174. doi: 10.1038/220173a0. [DOI] [PubMed] [Google Scholar]


