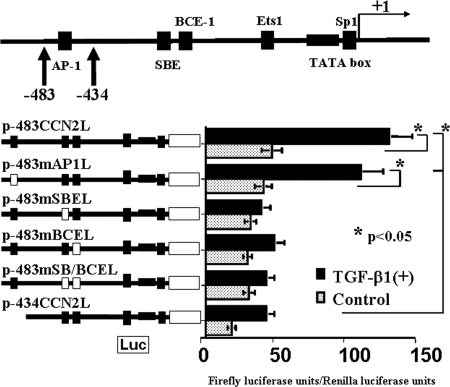
Figure 2.
Analysis of positive cis-regulatory elements at −483 to −434 bp within the CCN2 promoter. A putative AP-1 site is identified in this region, and there are known positive regulatory elements such as SBE, BCE-1, Ets1, and SP-1 sites within a region corresponding to −434 to 1 bp within the CCN2 promoter. The plasmids p-483mAP1L, p-483mSBEL, p-483mBCEL, and p-483mSB/BCEL contained mutated consensus motifs in their genomic sequences as shown in Table 1. Mutation of the AP-1 site was without effects so that other active elements would seem to be present within the region corresponding to −483 to −434 bp (p-483CCN2L versus p-483mAP1L). All of the regulatory elements (i.e., ones within a region corresponding to −483 to −434 bp and the SBE and BCE-1) were necessary for the full activity of the CCN2 promoter in response to rhTGF-β1 in mProx24 cells (p-483CCN2L versus p-483mSBEL, p-483mBCEL, p-483mSB/BCEL, and p-434CCN2L). The data were obtained from four independent experiments.